I'm trying to obtain the variance-covariance matrix of a logistic regression:
mydata <- read.csv("https://stats.idre.ucla.edu/stat/data/binary.csv")
mylogit <- glm(admit ~ gre + gpa, data = mydata, family = "binomial")
through matrix computation. I have been following the example published here for the basic linear regression
X <- as.matrix(cbind(1, mydata[,c('gre','gpa')]))
beta.hat <- as.matrix(coef(mylogit))
Y <- as.matrix(mydata$admit)
y.hat <- X %*% beta.hat
n <- nrow(X)
p <- ncol(X)
sigma2 <- sum((Y - y.hat)^2)/(n - p)
v <- solve(t(X) %*% X) * sigma2
But then my var/cov matrix doesn't not equals the matrix computed with vcov()
v == vcov(mylogit)
1 gre gpa
1 FALSE FALSE FALSE
gre FALSE FALSE FALSE
gpa FALSE FALSE FALSE
Did I miss some log transformation?
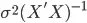
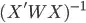
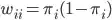
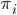 is the probability of event=1 at the observation level
is the probability of event=1 at the observation level