Bagging a RF model do not normally improve prediction performance(AUC) as RF already is bagged. If it does, probably some parameters in RF training are set suboptimal. So the easy answer is: don't bag the randomForest algorithm.
Also bagging RF could be computationally slow.
Bagging CART is a good idea. Infact so good, that some guys did just that plus some extra features and called it e.g. "random forests"! Bagging CART models is highly related to RF, but still inferior as mtry is set to the number of variables. This will lead to a little less decorrelated trees and lower robustness.
Bagging glm is a good idea for sparse and/or noisy datasets, as it will reduce overfitting. If you have a rich dataset 1000 samples and 10 features, don't bother as the unbiased glm will be very stable. Regularization could also be achieved more elegantly with the ridge-regression, elasticnet, lasso, PLS etc.. Check out glmnet package.
If two classifiers would to be directly combined, the voting is either unanimously or equal. This is of course a problem. Instead the classifiers can each be bagged(50 times e.g.) and all votes could be combined. This works fine, unless the dataset is unbalanced.
Many classifiers actual calculate some kind of probability measure and make decision here upon. This is true for logistic regression. For random forest the voting ratio can be understood as a pseudo probability. Remember to either stratify(down sampling) or use a lower weighting for the over-represented class. This will likely give a better model, see this answer and this answer on how to implement. If model do not provide any probability measure (regular SVM) it would be needed to bag the model to achieve a better fusing of models. In case of SVM it might smart to lower regularization to achieve more differentiated votes. As if the model is completely consistent, bagging will only provide the same result many times.
If one classifier is strongly superior, combining models is not likely to improve prediction performance. I tried to design a data-set of a linear component distributed on all variables (regression is good at this) and a non-linear component(RF can fit this) such that combining models is expected to improve performance. In example the models must identify if the sample is "spam" but any real text-analysis is left out. Because I could not make your github script work at first try.
I use a training-set to train, a calibration-set to decide weighting and a large test-set to measure performance. For real data, I would replace calibration and test set with nested n-fold cross-validation. But the example is already 100 lines, so I skip this for now.
Because you asked for bagging, I regularized glm by bagging and not by any other method. Otherwise I would have used elastic-net and RF and combined the probabilistic predictions directly.
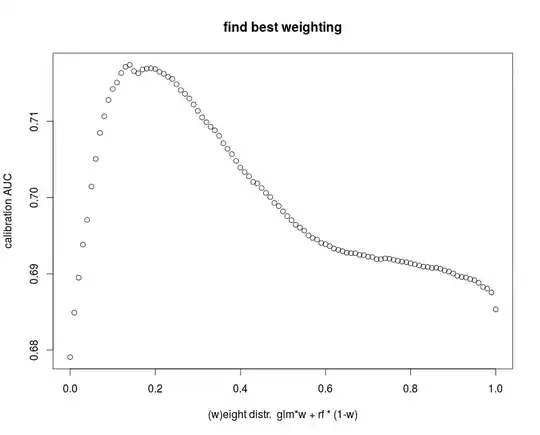
I have tried to write all bagging function so general that you easily can use them for your project even with completely different models. There is a makeSimData, trainBag, predictBag and a combine function for glm. I train a RF model and bagged glm. I use the probabilistic voting of either models and find best weighting with calibration-set. Other parameters could be tuned also. Again, model weights and model parameters could also be found directly by cross validation on training set.
To speed things up a little bagging can easily be parallelized. But this implementation will not work in windows, if running in windows set parallel=FALSE.
Lastly I post roc-AUC on test set. Of either RF and bagged-glm alone or in combination with the best found linear weighting rule.
and lastly the code:
rm(list=ls())
library(parallel) #for linux&mac only
#for windows use parallel=F below
library(randomForest)
library(AUC)
std = function(x) x/sd(x) #scaling function
makeSimData = function(
N = 500,
var = 250,
linW = .7,
noiseW = 1,
spamRatio = .5,
hidden.function = function(X) {
y.linear = apply(X,1,sum) #something glm is best at
y.quadratic = X[,1]^2 #something glm cannot fit
y.value = std(y.linear) * linW +
std(y.quadratic) * (1-linW) +
rnorm(N) * noiseW * 2 #noise component
y.class = (y.value<=quantile(y.value,spamRatio))+1
}
) {
X = replicate(var,rnorm(N))
y = c("regular","spam")[hidden.function(X)]
Data = data.frame(y=y,X=X)
}
trainBag = function(
Data, #defined data structure as makeSimData
model_func, #defined model function
trainPars, #what we
reps=11,
sampsizeRatio= 1, #
parallel=TRUE) {
if(parallel) nCore=detectCores() else nCore=1
Nsamples = dim(Data)[1]
sampsize = ceiling(sampsizeRatio * Nsamples)
preds = mclapply(1:reps,function(r) {
bootstrapData = Data[sample(Nsamples,sampsize,replace=T),]
do.call(model_func,trainPars)
},mc.cores=nCore)
}
predictBag = function(
myBag,
newData,
predPars,
combine = c,
parallel=TRUE
) {
if(parallel) nCore=detectCores() else nCore=1
preds = mclapply(myBag, function(model.fit) do.call(predict,predPars),
mc.cores=nCore)
out = combine(preds)
}
#make data
trainData = makeSimData(500,spamRatio=.5,noiseW=.2)
calibData = makeSimData(500,spamRatio=.5,noiseW=.2) #could also have used 10fold-CV
testData = makeSimData(5000,spamRatio=.5,noiseW=.2)
#train glm.bag and rf
glm.trainPars = alist(formula=y~., #define how glm should be run
data = bootstrapData,
family = "binomial")
glm.fit = do.call(glm,alist(formula=y~.,data=trainData,family="binomial"))
glm.bag = trainBag(trainData,glm,glm.trainPars)
rf.fit = randomForest(y~.,trainData)
#predict bag and rf
glm.predPars = alist(object = model.fit,
newdata=newData,
type="response")
#combine predicted probs, #could also be true/false ratio but that would stupid
glm_combine = function(bagPred) {
aMatrix = do.call(cbind,bagPred)
out = apply(aMatrix,1,mean) #combine by probabilistic average
#out = apply(aMatrix,1,x>0.5) #alternative combine by booleen voting
attributes(out) = NULL
return(out)
}
glmPred.calib = predictBag(glm.bag,
calibData,
glm.predPars,
glm_combine,
parallel=T)
rfPred.calib = predict(rf.fit,calibData,type="prob")[,2]
#tune weighting with calibration set (could also be some nFold-CV more lines...)
getBestWeight = function(Data,pred1,pred2) {
with(Data,{
weights = seq(0,1,le=101)
AUCbyWeight = sapply(weights, function(weight) {
auc( roc(pred1 * weight +
pred2 * (1-weight)
,y))
})
plot(
x=weights,
y=AUCbyWeight,
xlab = "(w)eight distr. glm*w + rf * (1-w)",
ylab = "calibration AUC",
main = "find best weighting",
)
#return best weight
weights[which(AUCbyWeight==max(AUCbyWeight))[1]]
})
}
#find best weight
bestWeight = getBestWeight(calibData,glmPred.calib,rfPred.calib)
#combine predictions with weighting, e.g. the best weighting
weightedPredict = function(Data,w,pred1,pred2,...) {
with(Data,{
weightedPred = pred1 * w + pred2 * (1-w)
plot(roc(pred1,y),add=F,col="green",...)
plot(roc(pred2,y),add=T,col="red")
plot(roc(weightedPred,y),add=T,col="blue")
return(weightedPred)
})
}
glmPred.test = predictBag(glm.bag,
testData,
glm.predPars,
glm_combine,
parallel=T)
rfPred.test = predict(rf.fit,testData,type="prob")[,2]
#apply
weightedPred.test = weightedPredict(testData,bestWeight,glmPred.test,rfPred.test,
main="RF(red), bagged-glm(green), combined(blue)")

