I am using parallel analysis (Horn 1965) to determine how many principal components I can extract from my data.
I can add more variables to my dataset, but I cannot add more cases (I know, that's weird, see below for some more context).
Presently, using a fixed number of cases (77), and relatively few variables (17), I can only be parallel-analysis-certain about 2 components (because only those parallel-analysis discounted eigenvalues are > 1), which isn't great for my purposes.
I am wondering whether it would make sense to add more variables to alleviate this problem, that is, whether that would allow me to retain more components, that pass muster under parallel analysis.
It seems obvious that the random data eigenvalues from a parallel analysis will grow as more variables are added; it becomes more likely that by random chance, two cases will be similar one some of the many variables. The question is: how fast do the random eigenvalues grow, as more variables are added. If they grow linearly, I'm screwed, and there is no point in adding more variables: as I add another variable, any improvement in (raw) eigenvalues will (probabilistically) be "eaten up" by an equal increase in necessary parallel-analysis adjustment. If they grow at a diminishing rate as you variables (is that a concave relationship then?!), there is hope: as you add more variables, only some, but not all of the possible increase in eigenvalues must be discounted away.
I have gone ahead and simulated this (very) crudely in R, and here's what I got:
library(paran)
library(reshape2)
library(ggplot2)
resparan <- matrix(data = NA, nrow = 400, ncol = 400)
for(i in 2:401) {
rdata <- matrix(data = rnorm(n = 77, mean = 0, sd = 2.8), nrow = 77, ncol = i)
resparan[1:i, i] <- paran(x = rdata, iterations = 100, centile = 95, quietly = TRUE, status = FALSE)$RndEv
print(paste("Calculating for", i, "variables."))
flush.console()
}
resparan.long <- melt(data = resparan, varnames = c("ncomps", "nvars"), value.name = "randomeigen")

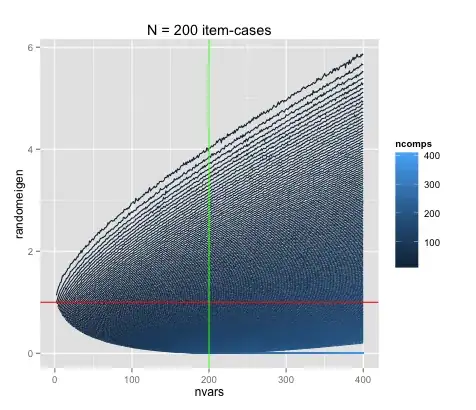
Above are two incarnations of the same plot, once for N (of item-cases) = 77, and once for N = 200.
The point where the number of cases equals the number of variables is denoted by green line.
The red line indicates Eigenvalue = 1, the Kaiser-Guttman criterion.
It appears that the curvature extends up to the point where the number of variables equals the number of cases – does that make sense?
Note: don't do this at home, takes about 20mins or so.
It seems, that in fact, the relationship is not linear, and there is hope.
I am guessing that aside from this numerical simulation, there must be math to bear this out. Correct? What would be a source or proof for the non-linearity of random data eigenvalue growth as more variables are added?
Clarifications:
- The actual, observed people-variables to be added will (hopefully) not be random. However, I am here not interested in the development of their unadjusted (or otherwise), observed eigenvalues: those are an empirical phenomenon. I want to know how eigenvalues from random data (which parallel analysis discounts by) develop, as more random variables are added. (That's what parallel analysis does: it runs PCA on many random datasets).
- By construction (see below), the additional random variables all have the same variance and mean.
Why would anyone want to do this? (Some context).
I'm glad you asked.
It's called Q Methodology (if anyone with sufficient privileges could add a new tag to that effect, that would be great).
In Q methodology, people as people-variables are asked to sort a (<100) number of statements as item-cases under a normal distribution.
(Because they must sort all items under a given (normal), forced distribution, the SD and mean of all Q-sorts as people-variables will be the same, in this case sd = 2.8 and mean = 0.)
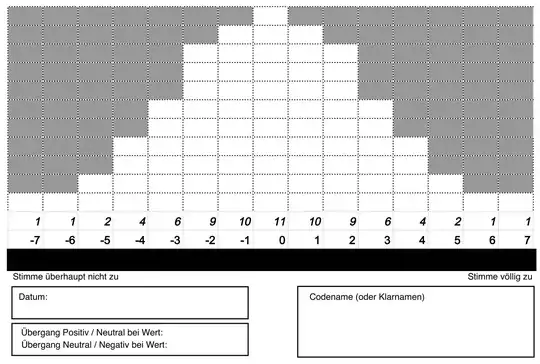
People place 77 cards with item-cases into the white boxes in the below template (unforunately in german).
*The x-axis values of any given item-case is that case's value on the person-variable of that Q-sorter.
You then extract factors (principal components, in my case) from the ranks of the cards, correlated across the people-variables. So it's basically like a normal factor analysis, but with a transposed data table. The idea is that the resulting factors (and their item-cases scores) can be interpreted as ideal-typical, shared viewpoints of people.
On the one hand, you want as many of such factors as possible, you decidedly don't care in Q by how many people any given viewpoint is shared. On the other hand, you do want to be certain that the viewpoint is really shared, and not just some random fluke, which you then interpret, making an ass of yourself in the process.
In this context, obviously, you cannot increase the number of item-cases above 100 or so, because people just can't be bothered with rank-ordering thousands of cards. You can, however, quite easily increase the number of variables, simply by having more people do the sorting.
Would that help?
I am quite fond of the rigorous standard of parallel analysis, I'm just not sure how it would translate to Q methodology.
Any and all suggestions and feedback would be very welcome, including explanations why my question is irrelevant / ill-formed, etc.
PS: it would be really nice if this could work. Not only would it allow a quite rigorous type of Q methodological work, it would also allow us to have some hunch about the necessary number of people-variables depending on the number of item-cases and desired number of components to be retained (if detected).