I have to solve a problem using a linear mixed model (lmer). Six subjects performed two tests, (test1, test2) in two different locations (on holiday, at work). These are my data:
subj <- c(1,1,2,2,3,3,4,4,5,5,6,6)
location <- c("holiday","holiday","holiday","holiday","holiday","holiday",
"work","work","work","work","work","work")
test <- c("test1", "test2","test1", "test2","test1", "test2","test1",
"test2","test1", "test2","test1", "test2")
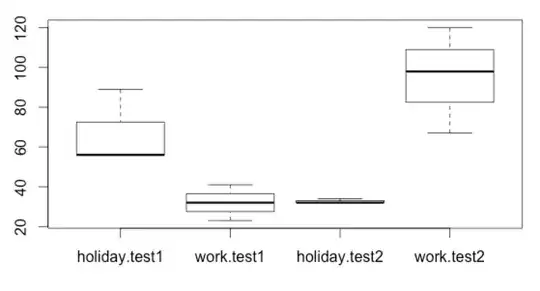
value <- c(56,32,89,32,56,34,23,98,32,120,41,67)
data <- data.frame(subj, location, test, value)
Using lmer, I have to verify if there is an effect of the variable location, if there is an effect of the variable test, and if there is an interaction, location X test.
I thought a model like this might be appropriate:
lmer(value ~ location*test + (1|subj))
Could it work?
Furthermore, I have to verify if there is a difference between holiday.test1 and work.test1, and between holiday.test2 and work.test2. I also need p-values.
`fm1 = lmer(value ~ test*location + (1|sub), data=tot)` and `fm2 = lmer(value ~ test+location + (1|sub), data=tot)` and using an `anova(fm1,fm2)`. In this case I could say: there is(or not) an interaction `test X location`. Then I could creating two models:`fm3 = lmer(value ~ test+location + (1|sub), data=tot)` and `fm4 = lmer(value ~ test + (1|sub), data=tot)` and verify if `location` affects value... it's correct? I'm a bit confused:) – Daryl Mar 31 '15 at 11:52