A lot of microbiome (microbial ecology) papers that I have come across use either principal component analysis (PCA) or principal coordinate analysis (PCoA) to make conclusions about the data. A lot of these claims are based upon components/coordinates with low variance, or by using the higher components to show patterns that aren't visible in the lower principle components. Although I've found a few questions here that cover the interpretation of PCA/PCoA plots, I haven't found any discussion of whether or not meaningful inference can be made based on components that explain lower variance.
The plot below is from a paper that compared the gut bacteria in pregnant women at different times. Each of the points represents a bacterial community. It does seem like the T1 samples are clustered together on the left of the figure, but is this meaningful when the component variances are only 8.9% and 4.5% respectively?
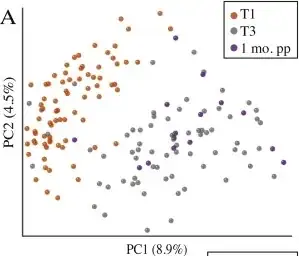
My second question is if it makes sense to make inferences based on patterns visible in the higher components, when these patterns aren't visible in the lower ones.
A good example of this is from the Human Microbiome coursera course. The plot below shows how the bacterial communities cluster from different body parts. In this example, the vaginal communities (shown in purple) cluster with those from the skin (shown in green).
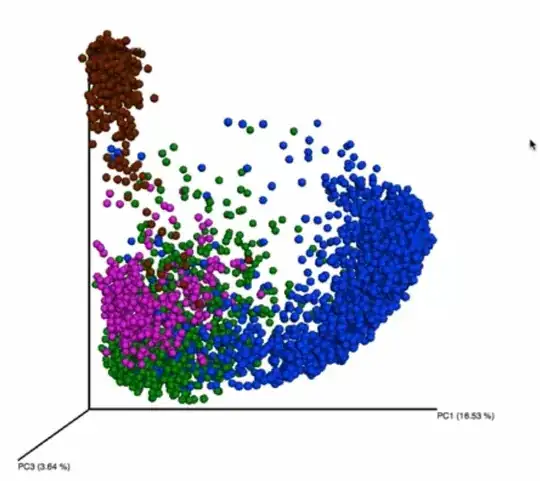
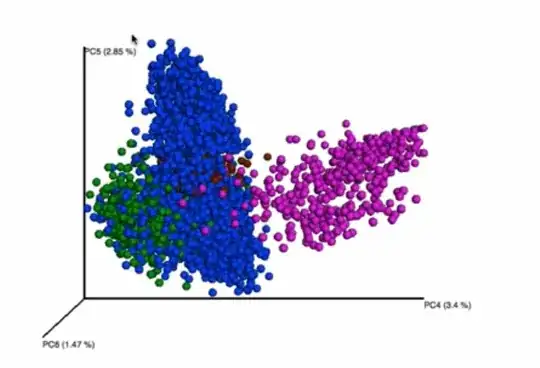
However, this community seems to cluster by itself when you only look at principal components four through six. Is it an acceptable practice continue looking at the other components, when you aren't getting separate clusters in first few? To me this feels like you are fishing for the results that you want to see.
I would greatly appreciate any insight about this topic! Here are the related topics that I found here that didn't quite answer my question:
- Making sense of principal component analysis, eigenvectors & eigenvalues
- Interpreting Principal Component Analysis output
Sources: