I have two questions about output of hierarchical clustering and improving the output.
I'm trying to learn more about performing hierarchical clustering in R so I started looking at a simple dataset I created of sushi rolls at a local restaurant. I went though every roll on the menu and created a distinct list of the union of all ingredients.
Then for each roll I put a 1 if it had that particular ingredient or a 0 for not having it. I then calculated distance based on Jaccard distance and created a dendogram using four different methods.
So my first question is how to interpret these correctly. Since many rolls overlap on quite a bit of ingredients, single-linkage is producing clusters that aren't significantly different from each other? Complete-linkage and the two methods are better reflecting the diversity in the rolls where some are significantly different? I'm not sure I really understand the subtleties between these four methods.
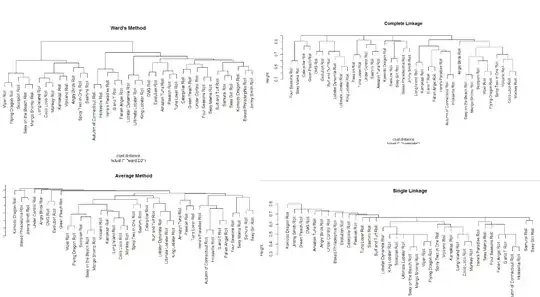
My second question is how would one handle related attributes? Say for example there exists {Tuna, White Tuna, Spicy Tuna} as three different ingredients. I think we would agree that the distance between {Tuna,Avocado} & {Salmon, Avocado} should be larger than {Tuna, Avocado} & {Spicy Tuna, Avocado}. Are the attributes typically collapsed into {Tuna} or is there another way to reflect the relationship?