Animal model (frequently used in animal science and sometime in human or plants) is mixed model with:
$y$ = $X$$b$ + $Z$$u$ + $e$
y is observed values for any quantitative variable, $Xb$ is fixed effects terms while $Zu$ is random effects, and $e$ is residual. Covariance between terms of $u$ can be defined as: $G$ = $A$$\sigma$$^2$$u$ . $A$ is additive relationship matrix whereas $\sigma$$^2$$u$ is additive genetic variance. $A$ can estimated from pedigree or from markers most commonly as $M$$M'$ where $M$ is marker scores (-1,0,1) x observation matrix.
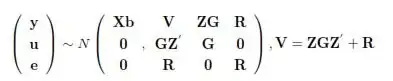
If there are no fixed effects in the model above model can be re-written as:
$y$ = $Z$$u$ + $e$
As $A$ is estimated from large number of markers with multi-colinarity it is logical to shrink the $u$ toward 0 using ridge shrinkage. From the above information we can calculate Best Linear Predictors (BLUPs).

Here for individual i, the BLUP estimate is $yi$, which is summation of $ai$ is additive genetic effect and $ei$ is residual.
Is there any R software package that can do this ? Examples appreciated.