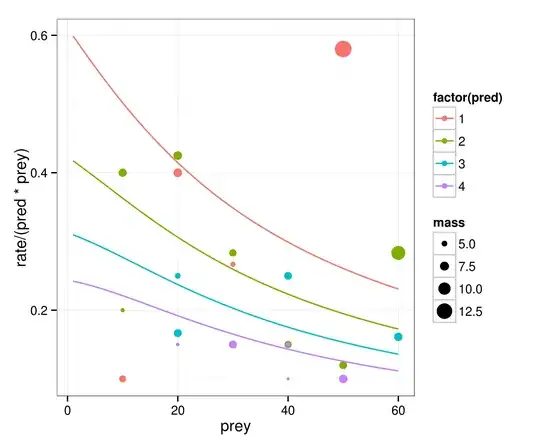
I am trying to fit several MLEs on a small dataset. I have managed to get it done for all my potential models except for this last one. This last one is actually the model that I expected to be the best model, because it was the best model when I did the whole thing using GLS. To start, here is the data:
dat$rate <- c(8,1,17,34,8,30,8,15,17,4,29,12,29,12,12,18,24,20,10,2,4)
dat$pred <- c(2,1,2,2,1,3,1,3,2,1,1,2,3,2,4,4,4,4,3,1,2)
dat$prey <- c(10,10,20,60,20,40,30,20,30,40,50,40,60,50,20,30,40,50,20,10,10)
dat$mass <- c(6.885000,5.942000,7.199000,11.285500,7.263000,6.571667, 4.583000,4.949000,6.220000,2.730000,13.330000,6.074000,7.053667,6.626000, 3.291500,6.711750,4.530750,7.076750,6.644333,2.861000,3.793500)
Here is the function that contains the equation:
CM = function(N0,a,h,tt,P,m, S, al) {
a = a/(1+m*(P-1)+a*h*m*(P-1)*N0)
h=h*S^al
N0 - lambertW(a*h*N0*exp(-a*(P*tt-h*N0)))/(a*h)
}
And here the model:
model = function(eaten, initial, attack, handling, time.exp, pred.no, interf, mass, allometric) {
if (attack < 0 || handling < 0) return(NA)
prop.exp = CM(N0=initial, a = attack, h = handling, tt = time.exp, P = pred.no, m=interf, S= mass, al= allometric)/initial
likelihoods = dbinom(eaten, prob = prop.exp, size = initial, log = TRUE)
negLL = -sum(likelihoods)
return(negLL)
}
And finally the code to fit the model
cm3 = mle2(model,
start = list(attack = .02, handling = .02, interf=0.02, allometric = -0.2),
data = list(initial = dat$prey,
eaten = dat$rate,
pred.no = dat$pred,
mass = dat$mass
),
fixed = list(time.exp = 1)
)
I have tried many different starting values and they all ended up with error messages like:
non-finite finite-difference value [2]
or
initial value in 'vmmin' is not finite
I guess that either "attack" or "handling" returns an invalid value but I am not sure how to fix this. Any ideas or suggestion are really appreciated.
Thanks in advanced, Robbie
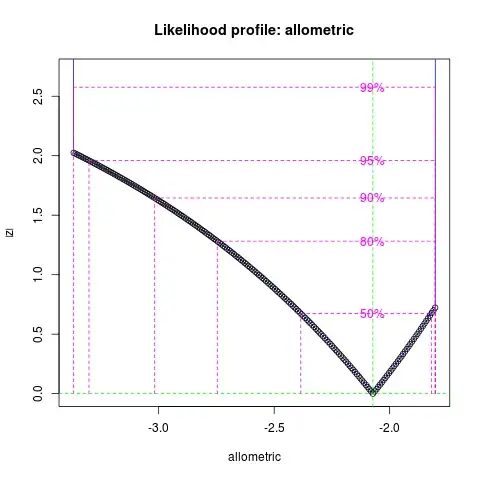
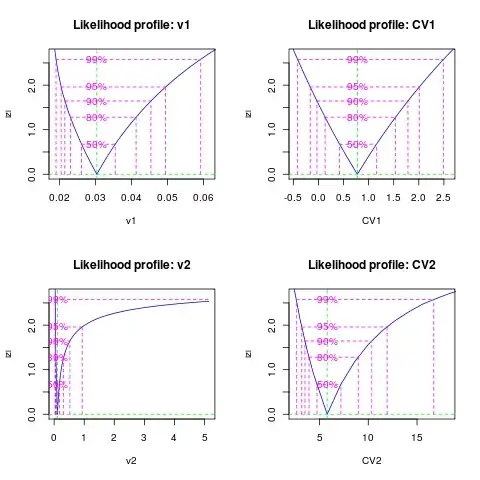 Confidence intervals:
Confidence intervals: