I've got data plotted for A/Ci curves (plant physiology, A=photosynthesis rate, Ci=internal CO2 concentration--essentially it's a dose-response relationship). Generally, when this type of data is presented, people just add SE error bars (like the provided image) and if they don't overlap call it significant, or they run an ANOVA on individual pairs of points (such as the two points, black and white, that sit above Ci=120).
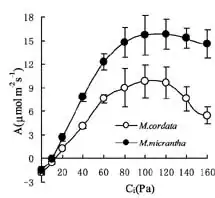
However, I would like to find a single test for the overall dose-response relationship. So, essentially I can assert that the dose-response relationship is significantly different between the two groups (in my case, the groups are all for the same species--it's a fertilizer experiment and I want to be able to claim that the addition of fertilizer changes the kinetics of the A/Ci relationship). I would also like to be able to test more than two curves on the same plot in the same manner (but I only have 3 curves, so I could do permutations if necessary).
Also, one other issue (not represented in this image) is that there is also an X-axis error term, so the error bars should actually be crosses, not just error on the y axis--which makes using an ANOVA approach difficult, since you have to test points in both dimensions.
Any suggestions for an appropriate test? I work mostly in R, so any link to existing scripts would be ideal--but I also have access to Graphpad Prism and JMP if anyone knows how to run this type of test in either of them.