You can play with this code: (taken and modify from: http://scikit-learn.org/stable/auto_examples/svm/plot_iris.html)
I cheated a bit to have only one feature but I guess you get the point.
Just think of it in 2D using only a line and not in 3D
import numpy as np
import pylab as pl
from sklearn import svm, datasets
def linearly_separable_data():
X = np.r_[ np.random.randn(20,2) - [3,3], np.random.randn(20,2) + [4,4]]
X[:,1] = 0
Y = [0]*20 + [1]*20
return X, Y
def non_linearly_separable_data():
X = np.r_[ np.random.randn(20,2) - [3,3], np.random.randn(20,2) + [2,2], np.random.randn(20,2) + [5,5]]
X[:,1] = 0
Y = [0]*20 + [1]*20 + [0]*20
return X, Y
X, Y = non_linearly_separable_data()
h = .02 # step size in the mesh
# we create an instance of SVM and fit out data. We do not scale our
# data since we want to plot the support vectors
C = 1.0 # SVM regularization parameter
svc = svm.SVC(kernel='linear', C=C).fit(X, Y)
rbf_svc = svm.SVC(kernel='rbf', gamma=0.7, C=C).fit(X, Y)
poly_svc = svm.SVC(kernel='poly', degree=3, C=C).fit(X, Y)
lin_svc = svm.LinearSVC(C=C).fit(X, Y)
# create a mesh to plot in
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
# title for the plots
titles = ['SVC with linear kernel',
'SVC with RBF kernel',
'SVC with polynomial (degree 3) kernel',
'LinearSVC (linear kernel)']
for i, clf in enumerate((svc, rbf_svc, poly_svc, lin_svc)):
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, m_max]x[y_min, y_max].
pl.subplot(2, 2, i + 1)
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape(xx.shape)
pl.contourf(xx, yy, Z, cmap=pl.cm.Paired)
pl.axis('off')
# Plot also the training points
pl.scatter(X[:, 0], X[:, 1], c=Y, cmap=pl.cm.Paired)
pl.title(titles[i])
pl.show()
For linearly separable:
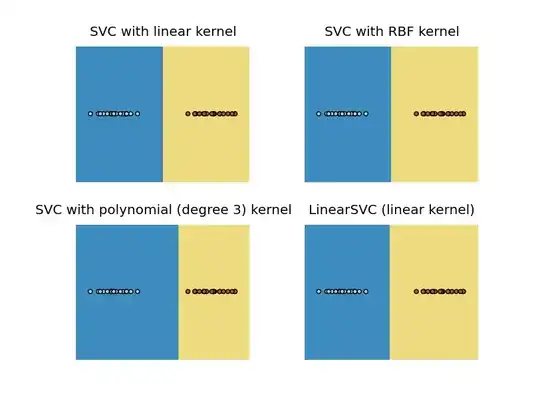
And for non-linearly separable:
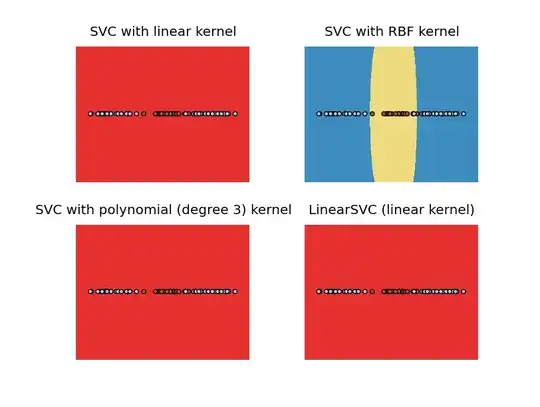
So for the linearly separable case, you got a point separating the two classes.
And for non-linearly separable, you got an interval.

