I am kind of stuck with an easy question:
I have two data sets with experimental data. The data sets do not have the same size. I would like to show that these data sets are possibly coming from the same experiment.
I tried a two-sample $t$-test; it shows that the data are significantly different. Is there a way to generate something like a $p$-value for similarity instead of difference?
Update:
Here an example:
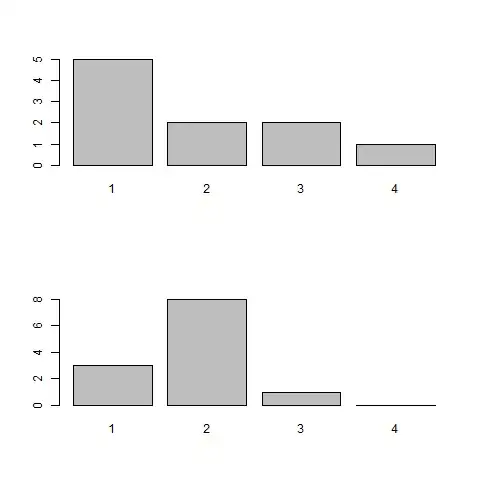
Date set 1 (Vector): 1 1 2 3 1 2 1 3 4 1 Mean: 1.9
Data set 2 (Vector): 2 2 1 2 2 1 1 2 2 3 2 2 Mean: 1.83
How would you now show that this data sets are from one experiment?