When including polynomials and interactions between them, multicollinearity can be a big problem; one approach is to look at orthogonal polynomials.
Generally, orthogonal polynomials are a family of polynomials which are orthogonal with
respect to some inner product.
So for example in the case of polynomials over some region with weight function $w$, the
inner product is $\int_a^bw(x)p_m(x)p_n(x)dx$ - orthogonality makes that inner product $0$
unless $m=n$.
The simplest example for continuous polynomials is the Legendre polynomials, which have
constant weight function over a finite real interval (commonly over $[-1,1]$).
In our case, the space (the observations themselves) is discrete, and our weight function is also constant (usually), so the orthogonal polynomials are a kind of discrete equivalent of Legendre polynomials. With the constant included in our predictors, the inner product is simply $p_m(x)^Tp_n(x) = \sum_i p_m(x_i)p_n(x_i)$.
For example, consider $x = 1,2,3,4,5$
Start with the constant column, $p_0(x) = x^0 = 1$. The next polynomial is of the form $ax-b$, but we're not worrying about scale at the moment, so $p_1(x) = x-\bar x = x-3$. The next polynomial would be of the form $ax^2+bx+c$; it turns out that $p_2(x)=(x-3)^2-2 = x^2-6x+7$ is orthogonal to the previous two:
x p0 p1 p2
1 1 -2 2
2 1 -1 -1
3 1 0 -2
4 1 1 -1
5 1 2 2
Frequently the basis is also normalized (producing an orthonormal family) - that is, the sums of squares of each term is set to be some constant (say, to $n$, or to $n-1$, so that the standard deviation is 1, or perhaps most frequently, to $1$).
Ways to orthogonalize a set of polynomial predictors include Gram-Schmidt orthogonalization, and Cholesky decomposition, though there are numerous other approaches.
Some of the advantages of orthogonal polynomials:
1) multicollinearity is a nonissue - these predictors are all orthogonal.
2) The low-order coefficients don't change as you add terms. If you fit a degree $k$ polynomial via orthogonal polynomials, you know the coefficients of a fit of all the lower order polynomials without re-fitting.
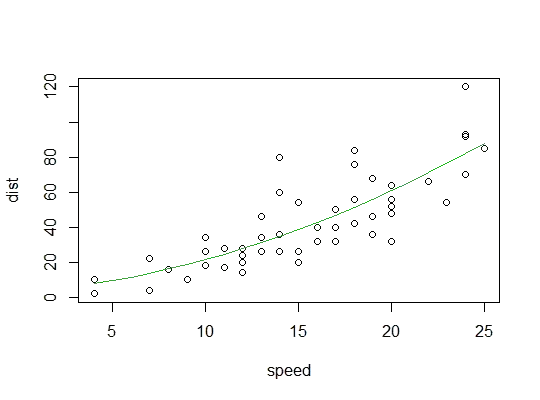
Example in R (cars data, stopping distances against speed):

Here we consider the possibility that a quadratic model might be suitable:
R uses the poly function to set up orthogonal polynomial predictors:
> p <- model.matrix(dist~poly(speed,2),cars)
> cbind(head(cars),head(p))
speed dist (Intercept) poly(speed, 2)1 poly(speed, 2)2
1 4 2 1 -0.3079956 0.41625480
2 4 10 1 -0.3079956 0.41625480
3 7 4 1 -0.2269442 0.16583013
4 7 22 1 -0.2269442 0.16583013
5 8 16 1 -0.1999270 0.09974267
6 9 10 1 -0.1729098 0.04234892
They're orthogonal:
> round(crossprod(p),9)
(Intercept) poly(speed, 2)1 poly(speed, 2)2
(Intercept) 50 0 0
poly(speed, 2)1 0 1 0
poly(speed, 2)2 0 0 1
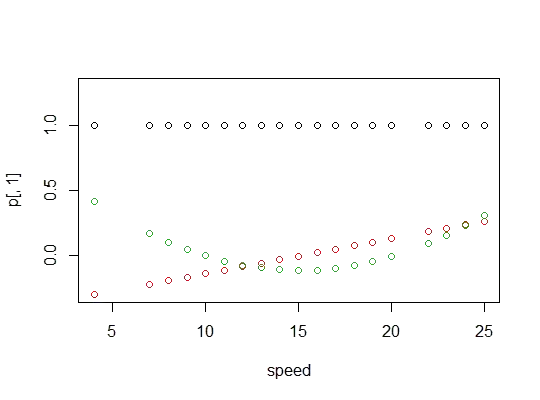
Here's a plot of the polynomials:
Here's the linear model output:
> summary(carsp)
Call:
lm(formula = dist ~ poly(speed, 2), data = cars)
Residuals:
Min 1Q Median 3Q Max
-28.720 -9.184 -3.188 4.628 45.152
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 42.980 2.146 20.026 < 2e-16 ***
poly(speed, 2)1 145.552 15.176 9.591 1.21e-12 ***
poly(speed, 2)2 22.996 15.176 1.515 0.136
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 15.18 on 47 degrees of freedom
Multiple R-squared: 0.6673, Adjusted R-squared: 0.6532
F-statistic: 47.14 on 2 and 47 DF, p-value: 5.852e-12
Here's a plot of the quadratic fit: