I ran lm() (I'm using R, but I don't think anything else in this question is R specific) then realized I'd used the wrong input data, basically meaningless. Yet, I got a really good model:
Residuals:
Min 1Q Median 3Q Max
-0.069654 -0.003899 -0.000381 0.004722 0.083622
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.795e-02 1.800e-03 9.970 <2e-16 ***
x -3.676e-06 3.848e-07 -9.554 <2e-16 ***
---
Residual standard error: 0.01416 on 11338 degrees of freedom
Multiple R-squared: 0.007986, Adjusted R-squared: 0.007899
F-statistic: 91.28 on 1 and 11338 DF, p-value: < 2.2e-16
I don't quite have permission to upload the data, and also it is large, but here is a plot of y and x (NB. x is on the vertical axis):
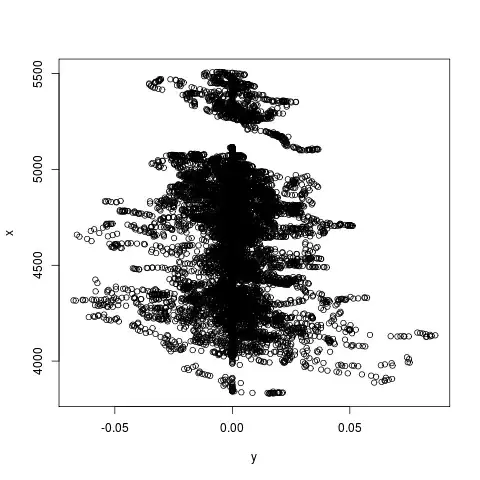
Okay, the break at roughly x==5200 is interesting, but basically it looks to me like for any value of x, y could be either side of y==0 with equal probability. So I don't see how it could possibly find a model with such a low p-value.
To help understand, I tried to do the same with random-looking data:
d=data.frame(
y=(rnorm(10000,0,0.1))^2,
x=5000+runif(10000)*500
)
d$y=ifelse(runif(10000)<0.5,-d$y,d$y)
But lm(y~x,data=d) gives the terrible model I expected:
Residuals:
Min 1Q Median 3Q Max
-0.165179 -0.004537 0.000104 0.004742 0.134604
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.439e-03 6.324e-03 0.228 0.820
x -2.967e-07 1.203e-06 -0.247 0.805
Residual standard error: 0.01737 on 9998 degrees of freedom
Multiple R-squared: 6.082e-06, Adjusted R-squared: -9.394e-05
F-statistic: 0.06081 on 1 and 9998 DF, p-value: 0.8052
That random data looks like this:
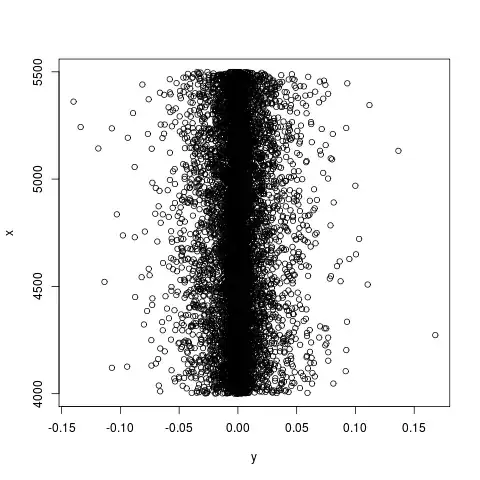
So, my real data and my random data behave completely differently when given to lm(), despite "x" looking equally useless as a predictor for "y" in both cases. It must be one of:
- I'm misreading the linear model output
- I'm asking the wrong question
- The "x" in my real data is genuinely a good predictor for "y"