I am currently trying to understand mixed effect models. And I would like to ask for some help understanding these results.
The data that I have is the mass volume of different rats across different days. Each rat has different time points where they took the measurement of that volume. There are 6 rats with volume measurements and 2 groups 3 rats came from Chile and 3 from England.
So the model is:
m1 <- lmer(lVolume ~ Country*Day + (1|Rat))
I am trying to understand how I can interpret the results from these plots:
***Update ***
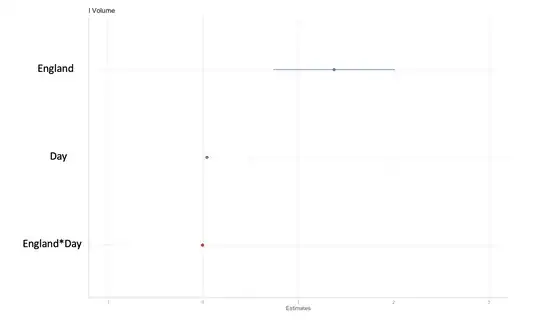
I plotted the estimates:
Fixed parameters betas:
coeff <- fixef(m1)[2] :
PassageChile
0.0458
(respectively for England*Day)
Random parameters :
coeff <- sqrt(VarCorr(m1)$Rat[1])
To see left plot (similarly for the right one just changes the coeff and conf_int):
ggplot(df, aes(x = Set_Rat_1, y = coeff, color = Country)) +
geom_point(show.legend = FALSE, size = 3,
position = position_dodge(0.5)) +
geom_errorbar(aes(x = Set_Rat_1, ymin = Lower_confidence,
ymax = Upper_confidence), width = 0.2,
position = position_dodge(0.5), show.legend = FALSE) +
scale_shape_manual(values = cols) +
labs(x = "Set_Rat_1", y = "FE", title = "Set Rat 1") +
theme_bw()
In addition, I got this but how do interpret these single points in terms of fixed effect, random effect and volume growth?
df.plot = ggpredict(model = m1,
terms = "Day",
type = "fe")
ggplot(data = df.plot,
mapping = aes(x = x,
y = exp(predicted),
ymin = exp(conf.low),
ymax = exp(conf.high))) +
geom_ribbon(fill = "lightblue") +
geom_line(size = 1)
plot_model(
m1,
bpe = "mean",
bpe.style = "dot",
prob.inner = .4,
prob.outer = .8
)
In that sense,
a) I would like to interpret these plots with the model, what this single value in each country is telling me for both the fixed effect and the random effect.
b) Is there a way to check the significance between these 2 points from the fixed effect and the 2 points from the random effect?
c) Also, when I include more rats and plot the predictions from the m1 model, you can kind of notice a clustering in the growth, is there a statistical way to check that or the lmer tells you about this in some of the parameters?
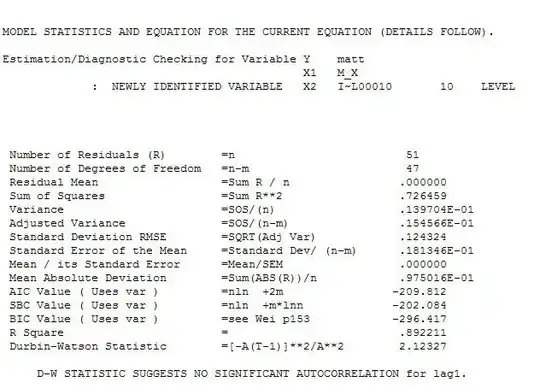
This is partial data that the model uses:
For rat 1 I have volume c(78, 304, 352, 690, 952, 1250) at days c(89, 110, 117, 124, 131, 138) that belong to country Chile.
For rat 2 I have volume c(202, 440, 520, 870, 1380) at days c(75, 89, 96, 103, 110) that belong to country Chile.
For rat 3 I have volume c(186, 370, 620, 850, 1150) at days c(75, 89, 96, 103, 110) that belong to country Chile.
For rat 4 I have volume c(92, 250, 430, 450, 510, 850, 1000, 1200) at days c(47, 61, 75, 82, 89, 97, 103, 110) that belong to country England.
For rat 5 I have volume c(110, 510, 710, 1200) at days c(47, 61, 75, 82) that belong to country England.
For rat 6 I have volume c(115, 380, 480, 540, 560, 850, 1150, 1350) at days c(47, 61, 75, 82, 89, 97, 103, 110) that belong to country England.