I am running a hierarchical Bayesian model using brms on reaction times (RTs) of a GoNogo task. The predictors are categorical and include the 3 stimuli/condition that participants observed and the 2 sequences of trials. I used an ex-Gaussian distribution for RTs.
here is the model:
informed1_RTs.bmodel = brm(RTs ~ conditionStimuli * sequenceTrials + (0 + conditionStimuli * sequenceTrials | Num_part)
, data = data_RTs_go
, family = exgaussian(link = "identity")
, warmup = 500
, iter = 2000
, chains = 2
, inits = "random"
, cores = 2
, seed = 123
, save_pars = save_pars(all = TRUE)
)
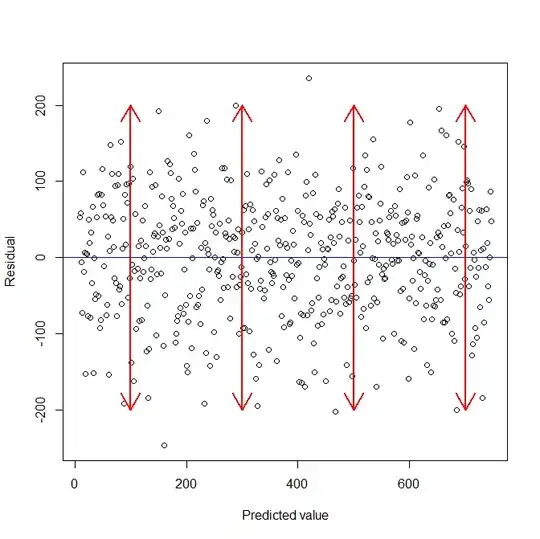
The pp_check shows a good fit (I think!) but the edges exceed the cut-off of the RTs (I chose a lower cutoff of 200ms and an upper cut-off o 750). I expect that inserting a boundary in the distribution reflecting the cut-off would improve the model predictions.
How do I insert such a boundary? Following Gelman, 2006 I tried to insert a uniform(200,750) but it gives various warnings and errors that the boundaries are not possible.
Thank you,
this is an example of the data:
Num_part trial_type Go_type conditionStimuli ITI_ms response RTs correctResponse order_pres sequenceTrials sdt
2 1 Go Bent leaves 819 1 301 1 1 NGG 1
3 1 Go Bent leaves 771 1 237 1 1 GG 1
4 1 Go Bent leaves 1086 1 393 1 1 GG 1
5 1 Go Straight leaves 652 1 331 1 1 GG 1
7 1 Go Bent leaves 919 1 372 1 1 NGG 1
9 1 Go Straight leaves 802 1 359 1 1 NGG 1