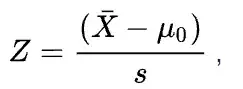How would one calculate the p-value from a doubly robust model
I could used the boot.pval r package and set the theta_null (The value of the parameter under the null hypothesis) 0, using student t-test as method.
Alternatively, I could use the approach below referencing: Computing p-value using bootstrap with R
How could one obtain the p-value for the doubly robust model? If by not by the approach below, then how?
time = c(14,18,11,13,18,17,21,9,16,17,14,15,
12,12,14,13,6,18,14,16,10,7,15,10)
group=c(rep(0:1, each=12))
sleep = data.frame(time, group)
require(boot)
diff = function(d1,i){
d = d1[i,]
Mean= tapply(X=d$time, INDEX=d$group, mean)
Diff = Mean[1]-Mean[2]
Diff
}
set.seed(1234)
b3 = boot(data = sleep, statistic = diff, R = 5000, strata=sleep$group)
b3.under.H0 <- b3$t - mean(b3$t)
pvalue = mean(abs(b3.under.H0) > abs(b3$t0))
pvalue
Updated: 2021-10-26:
My bootstrap distribution looks suitable for normal distribution but not t-distribution. When I used the percentile p-value calculation, the value 0.007 looked sensible. boot.pval does not seem to be giving sensible results p=1x10-16, nor does normal p-value.
Updated: 2021-10-26 17:09:
For Null hypothesis H0: μ=μ0 vs alternative hypothesis H1: μ≠μ0 , it is two-tailed.
which two-tailed p-values can be calculated as 2Φ(−|Z|) (for two-tailed tests) where Φ is the standard normal cumulative distribution function.