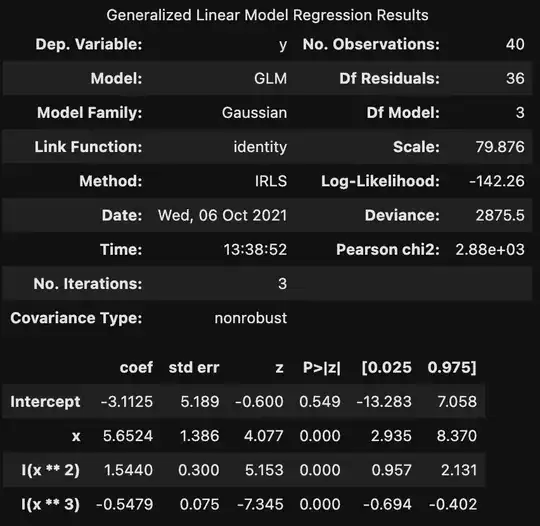
statsmodels library in python offers two different approaches for regression, ols and glm. While they are different methods, if the error is normally distributed, they should provide the exact same results (please see the code below).
However, one of the uses t-test and the other uses z-test. Consequently, the estimated confidence interval is different. We use t-test when sigma is not known and we estimate that based on the sample we have from the population. Can someone please explain why statsmodels.formula.api.glm uses z-test when family = sm.families.Gaussian()? Nowhere in the code we define sigma for the Gaussian distribution, so it is assumed to be unknown and we should estimate that using the sample observations we have. Considering that, we should use t-test then. Thank you
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import statsmodels.formula.api as smf
from statsmodels.stats.outliers_influence import summary_table
import scipy as sp
import math
from patsy import dmatrix
import scipy.stats
import statsmodels.api as sm
np.random.seed(seed=0)
Beg=-5
End=5
Samplingrate=40 #using more points, a more accurate spline can be generated
DrawingRate=100 #it is just for drawing the actual curve
#Data
#x
#xtrain=np.random.uniform(Beg,End,Samplingrate)
x=np.linspace(Beg,End,DrawingRate)
#y
#y=np.cos(x*np.pi/10 )
yactual=1+5*x+x**2-0.5*x**3
y=yactual+np.random.normal(loc=0.0, scale=10, size=DrawingRate)
#random selection
Selected=np.concatenate((np.ones(math.ceil(Samplingrate/2)),np.zeros(DrawingRate-Samplingrate),np.ones(math.floor(Samplingrate/2))),axis=None)
#np.random.shuffle(Selected)
#Traing Data
Data=pd.DataFrame({'x':x,'yactual':yactual,'y':y,'Selected':Selected})
Model2Ans=Data.copy()
Model3Ans=Data.copy()
DataTrain=Data[Data['Selected']==1]
DataTrain
#Model
DataTrain2=DataTrain.copy()
DataTrain3=DataTrain.copy()
Form='x+I(x**2)+I(x**3)'
model2 = smf.ols(formula = 'y ~ ' + Form, data = DataTrain).fit()
model3 = smf.glm(formula = 'y ~ ' + Form, data = DataTrain,family = sm.families.Gaussian()).fit()
DataTrain2['y_pred']=model2.predict(DataTrain2[['x','y']])
DataTrain2['y_error']=DataTrain2['y']-DataTrain2['y_pred']
DataTrain3['y_pred']=model3.predict(DataTrain3[['x','y']])
DataTrain3['y_error']=DataTrain3['y']-DataTrain3['y_pred']
print(model2.summary())
print(model3.summary())
predictions = model2.get_prediction(Data)
Model2Ans[['y_pred', 'mean_se', 'mean_ci_lower', 'mean_ci_upper', 'obs_ci_lower','obs_ci_upper']]= predictions.summary_frame(alpha=0.05)
Model2Ans
#for glm
predictions = model3.get_prediction(Data)
Model3Ans[['y_pred', 'mean_se', 'mean_ci_lower', 'mean_ci_upper']]= predictions.summary_frame(alpha=0.05)
Model3Ans
#Difference
Model2Ans[['y_pred', 'mean_se', 'mean_ci_lower', 'mean_ci_upper']]-Model3Ans[['y_pred', 'mean_se', 'mean_ci_lower', 'mean_ci_upper']]