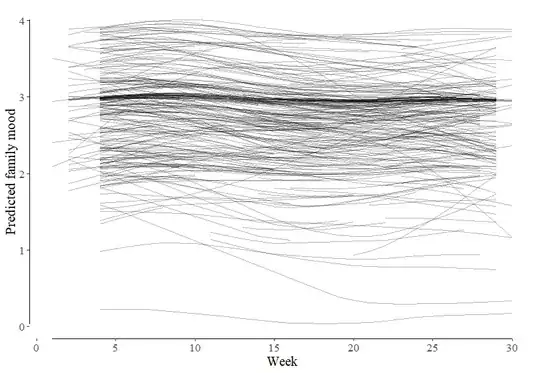
I have a problem with two forms of plots of two GAMM models: The first model is a baseline model estimating the time trend of measured mood during 30 weeks of 2020 (N=371). Lot's of missings, though (but I hope I can deal with that with a GAMM). The results for the first model show the best fit for a random smooth model:
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.69041 0.03282 81.97 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(x) 5.014 6.254 4.505 0.000118 ***
s(x,user_id) 550.871 1623.000 3.466 < 2e-16 ***
In the summary, x is the time variables (t = 1 to 30) and s(x,user_id) is the random smooth.
When I use the predict() function and plot the individual trends with ggplot, I get a picture of almost non-changing trends with a slight non-linearity (mood went down in fall 2020 and stayed there). However, some individuals started low and had a decline (5% of the sample).
The next progression in the model building is an interaction between time and self-reported health (mhem1_r) which turns out to be significant:
Approximate significance of smooth terms:
edf Ref.df F p-value
s(x) 5.068 6.317 4.365 0.000162 ***
s(x,user_id) 539.803 1623.000 2.830 < 2e-16 ***
s(mhem1_r) 1.827 1.876 51.953 < 2e-16 ***
ti(x,mhem1_r) 10.829 88.000 0.224 0.001788 **
When I use vis.gam() to get a 3D surface plot, three really weird things happen:
- The whole level of the surface is too low and differs from the baseline plot (which the intercepts in the output show).
- The trend looks so different: Whereas in the baseline plot, mood was rather stable, the vis.gam plot shows a downward trend followed by an upward trend.
- I really cannot see any interaction....
- A final thing: Although the main effect and interactions of/with perceived health are signficant, the R-sq is almost the same (.631 vs. .634) in both models.
Has anybody an idea what the problem is?
All the best and thank you in advance :)
Holger
P.S. Here is the code for both models in case this is the source of error
#Baseline model
gamm2c <- bam(mood ~ s(x, bs="cr", k=30) +
s(x, user_id, bs="fs", xt="cr", m=1, k=5)+
data=weekly_data, method="fREML")
#Interaction model
gamm_mhem <- bam(mood ~ s(x, bs="cr", k=30) +
s(x, user_id, bs="fs", xt="cr", m=1, k=5)+
s(mhem1_r, bs="cr", k=5) +
ti(x, mhem1_r, bs="cr", k = c(30,5)),
data=weekly_data, method="fREML")