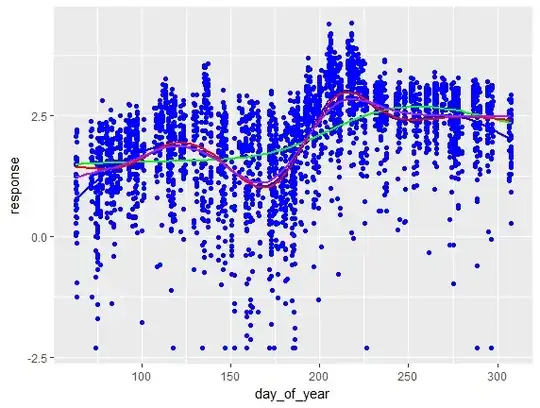
I am interested in the effect of dichotomous variable A on several scores for a sample size of N = 469 (observational data). Most scores have a non-normal, asymetrical distribution. Here's the contingency table when considering variables A and B, showing an unequal distribution, especially for the last level of variable B (n=96):
The strata/cells are uneven for the sample, although they are assumed to have an equivalent count in the general population.
More precisely, what I need is to know is what scores differ across the two levels of variable A when we control for variable B, then display descriptive statistics and graphs (e.g. boxplots) that show the difference for those scores. Ideally, scores should be kept on the original scale at least for the descriptives and graphs, since interpretation of the results depend heavily on the numerical values that are displayed.
Boxplots
Here is a group comparison for one of the scores, with the values taken directly from the sample (no bootstrap, no transformation). For that score, there was a difference in A for all levels of B.
For this other score, there was a difference in A only for some levels of B, notice also the inconsistent the shape of the distribution by levels of B:
Generalized linear model methodology
The original plan was to identify the scores that are different between the two levels of variable A across all levels of variable B by using a series of two-way ANOVAs (one for each score), such as some_score ~ A * B. I would then look for scores for which the A main effect is statistically significant while the main effect of B and the interaction term are not significant.
This approach had a few problems: (1) variance between groups was uneven when checking with a Levene test, (2) most scores have asymetrical distributions, (3) descriptive statistics and graphs (e.g. boxplots) comparing the levels of variable A would still be biased by the unequal distribution of variable B.
Update: To identify scores of interest, I used the Scheirer-Ray-Hare test as a non-parametric equivalent of two-way ANOVA. I selected scores that had a main effect for A, no main effect for B. The few scores that had a statistically significant interaction effect between A and B I examined with another non-parametric alternative, the Brunner-Munzel test, and found unsurprisingly that the interaction happened when there was a difference in A only at some levels of B. Another thing that I tried is a modified two-sample K-S test, with similar results. That solved my problems (1) and (2). For the descriptive stats and boxplots (problem 3) there was still an overrepresentation of some strata/cells, so I used a bootstrap methodology (see update below).
Bootstrapped methodology
Next attempt was to do a bootstrap resampling to control for unequal distribution of confounding categorical variable B, then use a bootstrapped test of stochastic dominance (e.g. Reiczigel et al. 2005) to determine which scores differ across groups. But that raised the problem of how many observations to draw from each "cell" (combination of A and B). Here's some solutions I considered:
Draw n = 37 for each cell because it's the smallest frequency count. Although that methodological choice seems intuitive, I have not found any studies backing the "smallest frequency as subsample size" rationale.
Draw n = 37 for the fist level of variable A, and n = 40 for the second level (smallest freq. count for that level), but again I see no clear justification for it besides intuition and convenience.
A problem that I see with the first two solutions is that, for cells with a higher count, variance is introduced by the larger pool of observations to draw from and by the sampling with repetition, while for cells with a smaller count, variance is only introduced by sampling with repetition.
Draw n = 47 from each subgroup so that the total bootstrap sample size is equal to the original sample size, but then some cells would get upsampled, other downsampled and this may be a problem.
A blend of solution 2 and 3: draw n = 41 for each level of variable B at the first level of variable A (to match the freq. count for that level), and n = 54 for each level of variable B for the second level of variable A.
Draw less than the smallest strata count (< 37), without replacement, i.e. repeated stratified sampling.
Inverse probability sampling (see update below).
Considering that the goal is to simulate a population where B is distributed evenly, is there a robust and reliable way to determine how many observations should be drawn from each cell ?
Update: I used inverse probability sampling, suggested in Nahorniak et al.. This creates subgroups that are even (on average) with less bias than stratified bootstrap (see proposed bootstrap method 3 above). The bootstrap results were used to describe group differences for scores that had been selected.