I have Per Capita fecundity of females from two population of Drosophila (evolved and ancestral-Population type) females at 5 different age points (age fixed factor), where same females were used for fecundity measurement. My unit of analysis for per capita fecundity at each age point(1,5,10,15 and20) comes by-counting the number of eggs laid by group of 10 females divided by number of females alive at the start of that day point. So basically its a fraction type data. This experiment was carried out with 5 independent replicate population of the two population type. Thus represents repeated measure (female fecundity measured at different age points)block design with 5 statistical blocks.
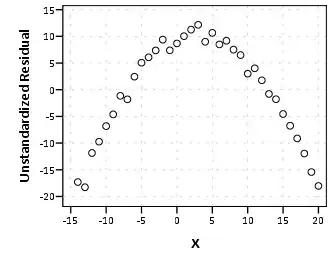
However, I got significant block effect and block interactions with other fixed effects, when I ran LMM under lme4 package (lmer) taking block as random factor. So we analysed each block seperately,therefore first checked for normality distribution of each block, 2 out of 5 blocks were not normally distributed (residual distribution was checked S-W test). Here's the qq-plot from one of those blocks (Block 1-W = 0.97244, p-value = 0.005062):
Other blocks showed better qq-plots, although the Shapiro-Wilk test still suggested non-normality (Block 5-W = 0.9795, p-value = 0.02518):
So whether can i still go for parametric test with this much deviation from normality can be accepted or should i go for non-parametric test for these 2 blocks. I thought of doing GLMM (glmer) but I am not aware of what distribution would fit my data type? Is it Poisson or quasipoisson or Gamma?