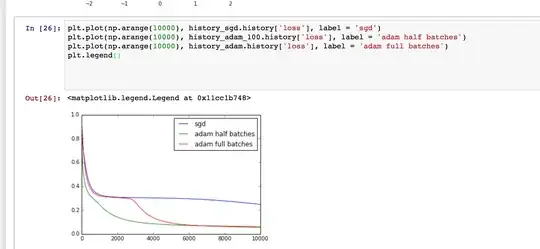
First, you hardly need a formal test with $n = 50\,000$ observations
and such an obviously bimodal histogram.
There are various kinds of tests of normality.
- The Shapiro-Wilk test, as implemented in R by
shapiro.test, will take up to 5000 observations.
- The Kolmogorov-Smirnov test, as implemented in R by
ks.test, will
test whether a sample follows a particular normal distribution. It would need to be modified, if $\mu$ and $\sigma$ were estimated from data.
- A chi-squared test based on $k$ histogram bins, with $\mu$ and $\sigma$ estimated from the binned data, would have $k - 3$ degrees of freedom. Results might vary according how the data are binned to make the histogram.
Each of these tests would require some modification or pre-processing before use.
Because I don't have access to the data in your histogram, I will use
somewhat similar fictitious data to illustrate a couple of possibilities.
set.seed(2021)
b = rbinom(50000, 1, .6)
x = rnorm(50000, 20.7, 1)
y = rnorm(50000, 24.2, 1.1)
z = b*x + (1-b)*y
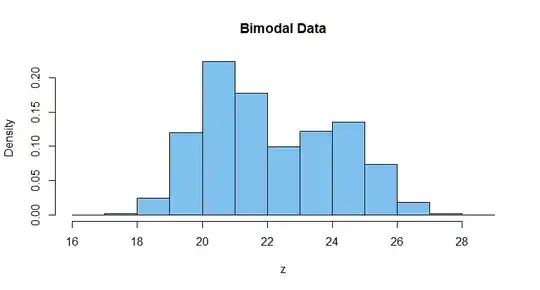
hist(z, prob=T, col="skyblue2", main="Bimodal Data")
One could sample $5000$ observations from among the $50\,000$ in z to
use shapiro.test, to get an unequivocal rejection of the null hypothesis that data are normal, with a P-value very near $0.$
shapiro.test(sample(z, 5000))
Shapiro-Wilk normality test
data: sample(z, 5000)
W = 0.96129, p-value < 2.2e-16
I am not sure why shapiro.test is limited to 5000 observations.
Perhaps it has to do with memory allocation or running time. However,
it may make sense to limit a goodness-of-fit test in this way to
avoid rejection on account of inconsequential quirks of the data
that may have nothing to do with the purpose of testing for normality.