You're missing a really important caveat to the question: even if I pick 3 breakpoints, the location of those breakpoints will affect the model performance. So not only the number, but the location is important. I will assume for this problem that you have a prespecified list of possible breakpoints and the question is which ones to use and when.
Since increasing the number of breakpoints increases the overall number of model parameters, this is actually a model or covariate selection problem. BIC and cross-validation are very amenable to this purpose. Basically, set-up an iterative procedure as follows:
- Simulate a dataset using k-fold cross validation.
- Fit all possible break-point models in your fixed/finite list of possible models
- Calculate the BIC for each model.
- Repeat 1-3 and calculate averages and error bars for the specific models and choose the one with lowest BIC.
Example using cumulative link for probit regression with ordinally valued outcomes having quadratic trend.
library(MASS)
library(splines)
set.seed(1234)
x <- rnorm(1000)
y <- rnorm(1000, -2 + .3 * x - .3*x^2, 0.3)
cutpoint <- quantile(y, c(0, 0.1, 0.4, 0.7, 1))
ycut <- cut(y, cutpoint, include.lowest=T)
## example of model output
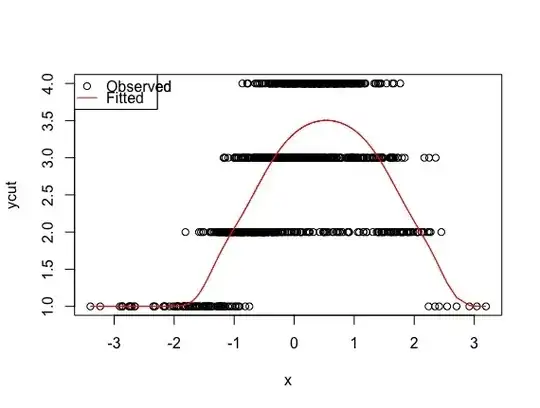
model <- polr(ycut ~ x + I(x^2), method='probit')
plot(x, ycut, main='Properly specified model')
predicted <- apply(predict(model, type='prob'), 1, weighted.mean, x=1:4)
lines(sort(x), predicted[order(x)], col='red')
legend('topleft', pch=c(1, NA), lty=c(0, 1), col=1:2, c('Observed', 'Fitted'))
## use no quadratic effects, expect to find breakpoint at apex
data <- data.frame('x'=x, 'y'=y, 'ycut'=ycut)
breakpoints <- c(-2, 0, 2)
ics <- replicate(1000, {
data <- data[sample(1:nrow(data), nrow(data)*0.75), ]
model1 <- polr(ycut ~ x, data=data, method='probit')
model2 <- polr(ycut ~ ns(x, df=1, knots=0), data=data, method='probit')
model3 <- polr(ycut ~ ns(x, df=1, knots=c(-1, 1)), data=data, method='probit')
model4 <- polr(ycut ~ ns(x, df=1, knots=c(-1, 0, 1)), data=data, method='probit')
ics <- c(BIC(model1), BIC(model2), BIC(model3), BIC(model4))
ics})
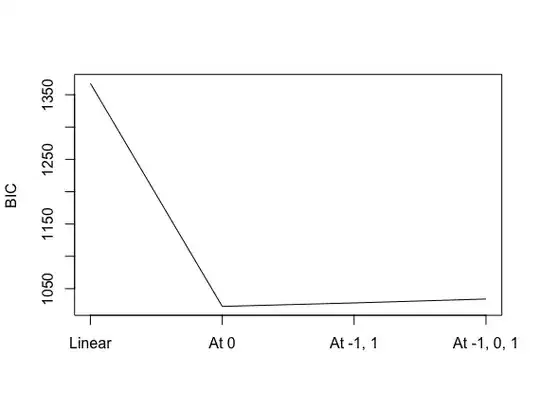
plot(rowMeans(ics), type='l', axes=F)
axis(1, at=1:4, labels=c('Linear', 'At 0', 'At -1, 1', 'At -1, 0, 1'))
plot(rowMeans(ics), type='l', axes=F, xlab='', ylab='BIC')
axis(1, at=1:4, labels=c('Linear', 'At 0', 'At -1, 1', 'At -1, 0, 1'))
axis(2)
box()
## best model, albeit the "wrong" one
best <- polr(formula = ycut ~ ns(x, df = 1, knots = 0), method = "probit")
plot(x, ycut, main='Properly specified model')
predicted <- apply(predict(best, type='prob'), 1, weighted.mean, x=1:4)
lines(sort(x), predicted[order(x)], col='red')
legend('topleft', pch=c(1, NA), lty=c(0, 1), col=1:2, c('Observed', 'Fitted'))
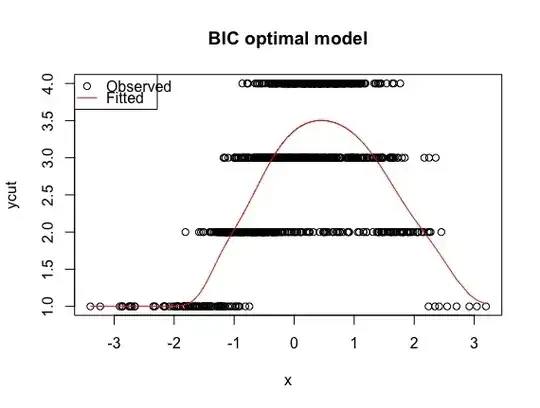
This method arrives at a near 100% agreement between the true model and BIC best model for predicting ordinal category based on the mode.
BIC optimal
Truth [-6.8,-3] (-3,-2.32] (-2.32,-1.96] (-1.96,-1.22]
[-6.8,-3] 81 2 0 0
(-3,-2.32] 1 284 1 0
(-2.32,-1.96] 0 1 209 16
(-1.96,-1.22] 0 0 10 395


