I am trying to jointly estimate the components of a mixture distribution.
I have a sampling from a mixture, XY, composed of X and Y with a known mixing parameter m. I also have a separate sampling of just Y. I am trying to estimate the the PDF of X.
Here is a concrete example.
# Generate XY sampling data
m <- 0.2 # Mixing parameter
n <- 1000
k <- rbinom(1, n, prob = m)
xy <- c(rnorm(n-k, 1, 1), rnorm(k, 5, 0.5))
# Estimate XY_pdf
XY_pdf <- density(xy)
plot(XY_pdf)
# Generate independent Y sampling data
y2 <- rnorm(500, mean = 5, sd = 0.5)
Y_pdf <- density(y2, bw = XY_pdf$bw)
lines(Y_pdf$x, Y_pdf$y*m, col = "red", lty=2)
# Function for calculating P_kde; https://stackoverflow.com/a/34682302/2723734
kde_val <- function(x, t, bw){
sapply(t, function(ti) {
kernelValues <- rep(0,length(x))
for(i in 1:length(x)){
transformed = (ti - x[i]) / bw
kernelValues[i] <- dnorm(transformed, mean = 0, sd = 1) / bw
}
return(sum(kernelValues) / length(x))
})
}
t <- seq(-3, 9, by = 0.01)
xy_val <- kde_val(xy, t, XY_pdf$bw)
y_val <- kde_val(y2, t, Y_pdf$bw) * m
x_val_est <- xy_val - y_val
lines(t, x_val_est, col = "blue")
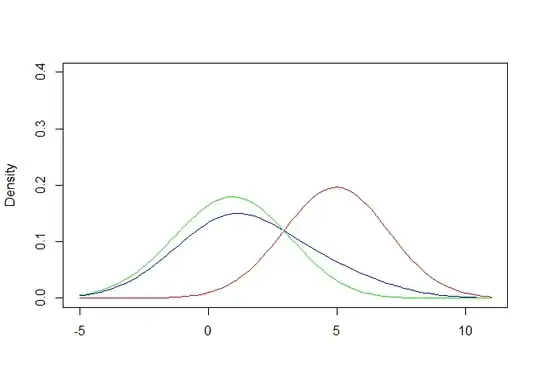
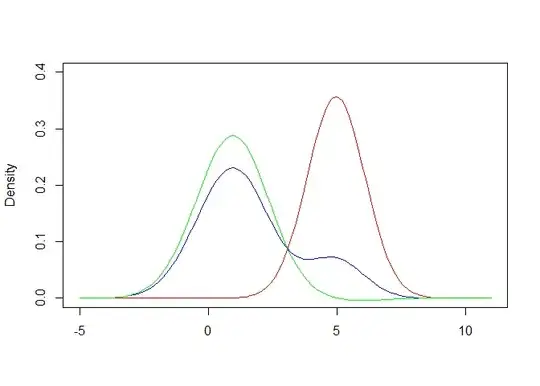
The plot shows PDF(XY) and PDF(Y) estimated from KDE (black and red); and an estimate of PDF(X) = PDF(XY) - PDF(Y)*m (blue).
The estimate of PDF(X) is pretty good, except towards the tail where it becomes negative due to sampling variation in XY and Y.
How do I properly estimate PDF(X)?
(I can't assume the distributions are gaussian)