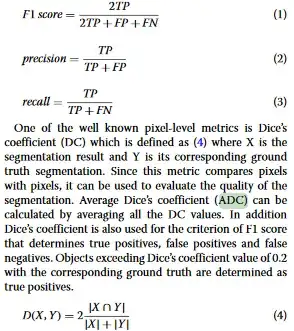
On page 8 of the paper An automatic nuclei segmentationmethod based on deep convolutional neuralnetworks for histopathology images, the authors show performance of their deep model on test sets. They have clearly mentioned two separate columns for F1 score and Average Dice Coefficient (ADC) (This answer does not answer my specific question):
Suppose the test set consisted of 4 images then Average Dice Coefficient (ADC) would be the average of Dice coefficient of 4 predictions of the input test images. They have defined F1 and Dice Coefficient as:
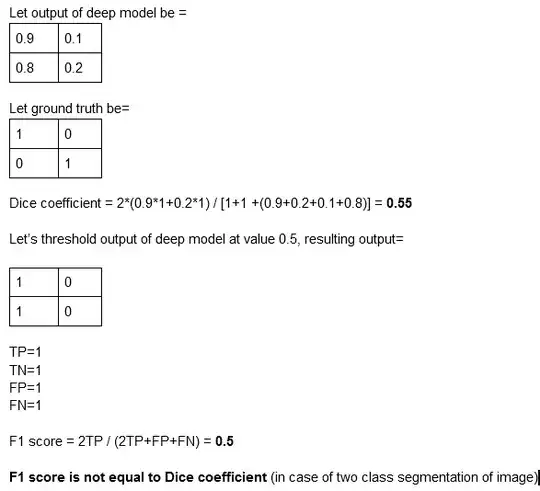
This is where my doubt comes. In binary segmentation (I mean two class segmentation, foreground or background), we get a sigmoid output. It consists of values in range [0,1], while ground truth contains only 1 and 0. Is it like for Dice coefficient, we take the sigmoid map as it is and compute Dice by (2*prediction*ground truth)/(prediction+ground truth) and for F1 we threshold sigmoid map so that it contains only 1 and 0 and then compute precision, recall and F1 by finding TPs, FPs and FNs? Is this the correct thinking or am I wrong?
To clarify my way of understanding here is an example:
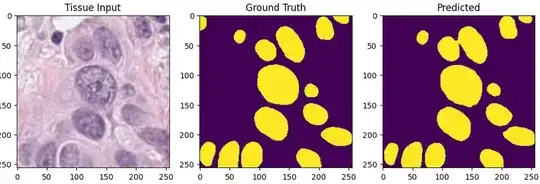
Example input image, ground truth and prediction, my example above considers 2x2 output and ground truth instead of 256x256 output and ground truth as in image below: