I have two distributions and I want to test whether there's an inequality of variances. They're non-normal, so Levene's test is appropriate. The scipy implementation offers three options: test on the mean, the median, and the trimmed mean. The trimmed mean is appropriate for heavy tailed distributions.
My question is, how do I know if my distribution is heavy-tailed? My understanding is that it's heavy-tailed if it's not exponentially bounded. I've tried to check this but I'm not sure if my method is correct. Here's what I did:
- Converted my data into z-scores so as to standardise it and plotted it.
Plotted the exponential distribution function across the range of my data, plotted it.
Compared the two visually.
EDIT: I'm here adding some descriptive statistics about my two distributions in response to the comments. This is the raw data, not the z scores.
Distribution 1:
count 38.000000
mean 1.160140
std 1.281058
min 0.220619
25% 0.451241
50% 0.623582
75% 1.478313
kurtosis 7.57
max 6.719054
Distribution 2:
count 40.000000
mean 0.887812
std 0.720215
min 0.252508
25% 0.408433
50% 0.617842
75% 1.120488
kurtosis 6.27
max 3.939130
My conclusion is that my distribution is not heavy tailed, but I'm not confident about it. Can anyone advise?
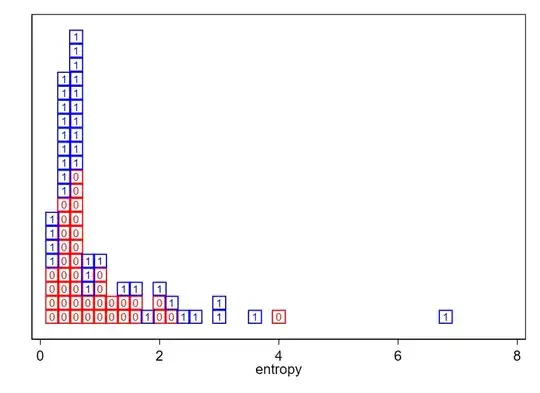
EDIT 2: This is my full data, long form:
Group entropy
1 6.71905356
1 0.56407487
1 0.738029138
1 0.630035416
1 3.017076375
1 2.510090903
1 0.254787047
1 0.376719953
1 0.456298101
1 0.328258469
1 0.767253283
1 0.641643213
1 2.905235741
1 3.615227362
1 0.244727319
1 2.317604878
1 1.504713298
1 0.999700392
1 0.669730607
1 0.766398132
1 0.449555621
1 0.360902977
1 0.297898424
1 1.399111031
1 0.67895411
1 0.56984134
1 0.536010552
1 2.226602414
1 1.998649951
1 0.220619041
1 0.547186366
1 0.446506256
1 0.495662791
1 0.458900635
0 1.699580285
0 1.017646859
0 0.618058775
0 0.740520854
0 0.558418925
0 0.264262271
0 1.4136416
0 0.538862166
0 2.089605078
0 2.206855803
0 0.494698728
0 0.36284015
0 0.947420619
0 1.515928283
0 0.682302263
0 0.515864165
0 0.400418084
0 0.401584527
0 1.195820577
0 0.544921866
0 0.284516915
0 1.902155181
0 1.095376897
0 0.263003363
0 0.674095659
0 3.939129819
0 0.617625765
0 0.364223021
0 0.355701427
0 0.887284165
0 0.312722361
0 0.570313528
0 0.4107156
0 0.453855313
0 1.441497841
1 1.720034593
1 0.590291826
1 0.444819008
0 0.252508237
0 1.226010557
0 0.526118886
0 1.046928619
0 0.679454156
1 0.617128565