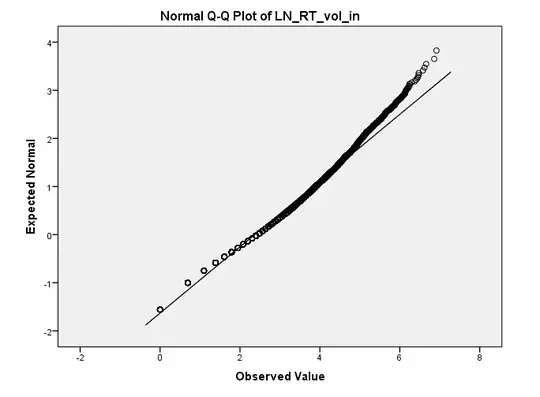
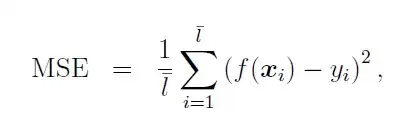You can plot this curve accurately, on a linear scale.
Let $a=1700$ and $b=300$ be the parameters.
The largest value of $f(x)=x^a(1-x)^b$ for $0\lt x \lt 1$ is attained at $x_m=(a-1)/(a+b-2)$ (the mode of the corresponding Beta$(a,b)$ distribution). There,
$$y_m = \log f(x_m) = a \log(x_m) + b \log(1-x_m)$$
is the logarithm of the curve's peak. Scale this up by defining
$$f^{*}(x) = \exp(-y_m) f(x) = \exp(a \log(x) + b \log(1-x) - y_m).$$
This attains the value of $1$ at its peak. Thus, the plot of $f^{*}$ fits within the vertical range $[0,1].$ To obtain a plot of $f,$ all you need do is relabel the vertical axis (by multiplying all its values by $\exp(y_m).$)
This approach works in any graphics environment. Here is an example, computed entirely with double-precision arithmetic in R:
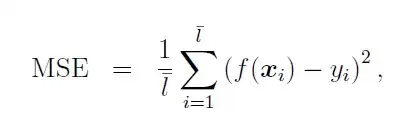
You can succeed provided both $a$ and $b$ do not themselves require more digits to represent than IEEE supports: that is, they should be a couple of orders of magnitude less than $10^{15.6}.$ Here is an example with $a=10^8$ and $b=10^{13}:$
(It is no accident both curves appear to have the same shape: both will be extremely close to a Gaussian when both $a$ and $b$ are large. For all practical purposes, all we ever have to do is draw a Gaussian and then label both axes suitably according to the parameters $a$ and $b$.)
This method will work whenever you can easily compute $\log f(x)$ and find (or estimate) its maximum value -- and this will be the case in most applications.
The R code below is merely to prove the concept: for general purpose work, the labeling algorithm will need to be a little fussier.
betaplot <- function(a, b, xlim=c(-4,4), scale=1, nticks=5, interval=2, ...) { # a,b>1
n <- a + b
mu <- a / n # Mean
sigma <- sqrt(((a / n) * (b / n)) / (n+1)) # SD
xlim <- xlim * sigma + mu # Plot limits
m <- (a - 1) / (n - 2) # Mode
f <- function(x) a * log10(x) + b * log10(1-x)
logmax <- round(f(m)) # Nearest whole power of 10 to max
#
# The plot itself.
#
curve(exp(log(10) * (f(x) - logmax)), xlim=xlim, ylim=c(0,scale), ylab="",
yaxt = "n", ...)
#
# Ticks and labels.
#
yticks <- seq(0, nticks) * interval
yticks <- yticks[yticks <= 10*scale]
rug(yticks/10, side=2, ticksize=-0.03)
for (y in yticks) {
if (y==0) {s <- 0} else
if (y==10) {s <- bquote(10^.(logmax))} else
{s <- bquote(.(y)%*%10^.(logmax-1))}
mtext(s, side=2, line=1, at=y/10)
}
}
#
# Examples.
#
betaplot(1700, 300, scale=0.8, nticks=4, interval=2, lwd=2)
betaplot(1e8, 1e13, scale=0.6, nticks=3, interval=2, lwd=2)