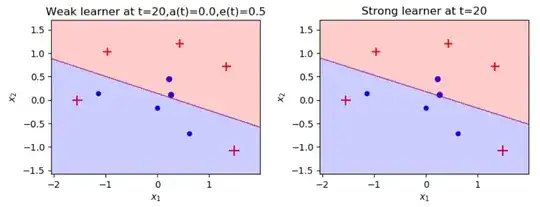
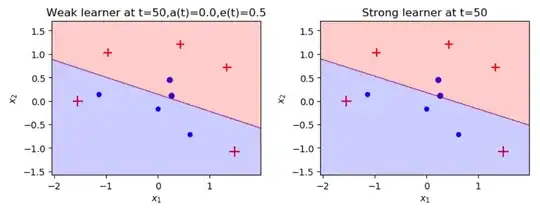
I implemented Adaboost that using weighted logistic regression instead of decision trees and I managed to get to 0.5% error, I'm trying to improve it for days with no success and I know it possible to get with him to 0% error, hope you guys could help me do it.
My logistic regression algorithm:
Lg.py:
import numpy as np
from scipy.optimize import fmin_tnc
class LogistReg:
def __init__(self,X,y,w):
self.X = np.c_[np.ones((X.shape[0],1)),X]
self.y = np.copy(y[:,np.newaxis])
self.y[self.y==-1]=0
self.theta = np.zeros((self.X.shape[1],1))
self.weights = w
def sigmoid(self, x):
return 1.0/(1.0 + np.exp(-x))
def net_input(self, theta, x):
return np.dot(x,theta)
def probability(self,theta, x):
return self.sigmoid(self.net_input(theta,x))
def cost_function(self,theta,x,y):
m = x.shape[0]
tmp = (y*np.log(self.probability(theta,x)) + (1-y)*np.log(1-self.probability(theta,x)))
total_cost = -(1.0/m )* np.sum(tmp*self.weights)/np.sum(self.weights)
return total_cost
def gradient(self,theta,x,y):
m = x.shape[0]
return (1.0/m)*np.dot(x.T,(self.sigmoid(self.net_input(theta,x))-y)*self.weights)
def fit(self):
opt_weights = fmin_tnc(func=self.cost_function,x0=self.theta,fprime=self.gradient,
args=(self.X,self.y.flatten()))
self.theta = opt_weights[0][:,np.newaxis]
return self
def predict(self,x):
tmp_x = np.c_[np.ones((x.shape[0],1)),x]
probs = self.probability(self.theta,tmp_x)
probs[probs<0.5] = -1
probs[probs>=0.5] = 1
return probs.squeeze()
def accuracy(self,x, actual_clases, probab_threshold = 0.5):
predicted_classes = (self.predict(x)>probab_threshold).astype(int)
predicted_classes = predicted_classes.flatten()
accuracy = np.mean(predicted_classes == actual_clases)
return accuracy*100.0
My Adaboost using WLR:
adaboost_lg.py:
import numpy as np
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
import matplotlib.pyplot as plt
from sklearn.datasets import make_gaussian_quantiles
from sklearn.model_selection import train_test_split
from plotting import plot_adaboost, plot_staged_adaboost
from Lg import LogistReg
class AdaBoostLg:
""" AdaBoost enemble classifier from scratch """
def __init__(self):
self.stumps = None
self.stump_weights = None
self.errors = None
self.sample_weights = None
def _check_X_y(self, X, y):
""" Validate assumptions about format of input data"""
assert set(y) == {-1, 1}, 'Response variable must be ±1'
return X, y
def fit(self, X: np.ndarray, y: np.ndarray, iters: int):
""" Fit the model using training data """
X, y = self._check_X_y(X, y)
n = X.shape[0]
# init numpy arrays
self.sample_weights = np.zeros(shape=(iters, n))
self.stumps = np.zeros(shape=iters, dtype=object)
self.stump_weights = np.zeros(shape=iters)
self.errors = np.zeros(shape=iters)
# initialize weights uniformly
self.sample_weights[0] = np.ones(shape=n) / n
for t in range(iters):
# fit weak learner
curr_sample_weights = self.sample_weights[t]
stump = LogistReg(X,y,curr_sample_weights)
#stump = LogisticRegression()
#stump = stump.fit(X, y, sample_weight=curr_sample_weights)
stump = stump.fit()
# calculate error and stump weight from weak learner prediction
stump_pred = stump.predict(X)
err = curr_sample_weights[(stump_pred != y)].sum()# / n
stump_weight = np.log((1 - err) / err) / 2
# update sample weights
new_sample_weights = (
curr_sample_weights * np.exp(-stump_weight * y * stump_pred)
)
new_sample_weights /= new_sample_weights.sum()
# If not final iteration, update sample weights for t+1
if t+1 < iters:
self.sample_weights[t+1] = new_sample_weights
# save results of iteration
self.stumps[t] = stump
self.stump_weights[t] = stump_weight
self.errors[t] = err
return self
def predict(self, X):
""" Make predictions using already fitted model """
stump_preds = np.array([stump.predict(X) for stump in self.stumps])
return np.sign(np.dot(self.stump_weights, stump_preds))
def make_toy_dataset(n: int = 100, random_seed: int = None):
""" Generate a toy dataset for evaluating AdaBoost classifiers """
n_per_class = int(n/2)
if random_seed:
np.random.seed(random_seed)
X, y = make_gaussian_quantiles(n_samples=n, n_features=2, n_classes=2)
return X, y*2-1
# assign our individually defined functions as methods of our classifier
if __name__ =='__main__':
X, y = make_toy_dataset(n=10, random_seed=10)
# y[y==-1] = 0
plot_adaboost(X, y)
clf = AdaBoostLg().fit(X, y, iters=20)
#plot_adaboost(X, y, clf)
train_err = (clf.predict(X) != y).mean()
#print(f'Train error: {train_err:.1%}')
plot_staged_adaboost(X, y, clf, 20)
plt.show()
It seems to me like the machine isn't learning in every regression iteration. I get the same result, even after the 50th iteration. I would like to know what I'm doing wrong, maybe my fit function not implemented well? or maybe my cost function?