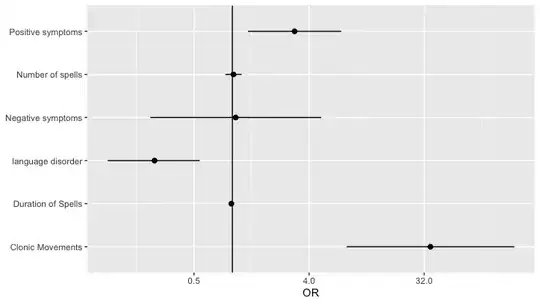
for a study i want to investigate which clinical symptoms of a patient, lead to the doctor stating a certain diagnosis (eg epilepsy). Ater using logistic regression to calculate the OR and 95% CI, I want to visualize the OR and 95% Ci in a forest plot. The problem however is that some symptoms have a very large OR and CI and some a small one, both being statistically and clinically significant. For example: clonic movements and language disorders (see below).
When plotted in a single figure (what i really want to do) the effect of language disorders is unreadable due to the large numbers of clonic movements. Any suggestion on how to visualize them in a different way? Tried using to take the natural log of all numbers, but then i come up with negative OR, which makes it even more unreadable?
Thanks in advance
Clinical symptom: OR + 95% CI
Number of spells 1.019 0.880- 1.181
Duration of Spells 0.982 0.966- 0.998
Positive symptoms 3.078 1.325- 7.151
Negative symptoms 1.062 0.227 – 4.980
Clonic Movements 35.8 7.876- 162.769
language disorder 0.245 0.105-0.553