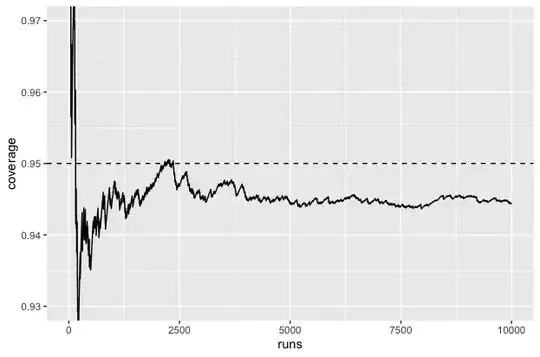
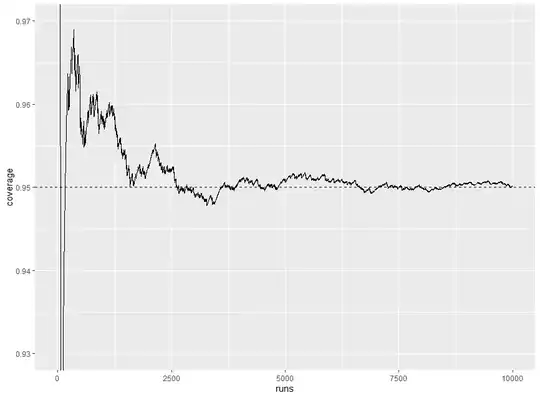
I have data that can be described using the model $y_{i j} = \alpha_i + \epsilon_{i j}$, where $\alpha_i \sim \text{N}(\mu_{\alpha}, \sigma_{\alpha}^2)$ and $\epsilon_{i j} \sim \text{N}(0, \sigma_{\epsilon}^2)$. There are 768 observations in 48 groups of 16 observations each, so $i = 1, \ldots, 48$ and $j = 1, \ldots, 16$. I want to calculate a 95% confidence interval for $\mu_{\alpha}$. I decided to try fitting the model $\tt{y \sim (1 \ | \ group)}$ to the data using the lme4 package and computing a confidence interval using the package's $\tt{confint()}$ function. I created a simulation to check the coverage of the confidence interval; the code is below. It seems like the coverage is around 94.5% instead of 95%. Is there a flaw in my simulation, or is this result to be expected?
# Load packages -----------------------------------------------------------
library(lme4)
library(tidyverse)
# Define functions --------------------------------------------------------
make_dataset <- function(groups, group_size,
mu_alpha, sigma_alpha, sigma_epsilon) {
replicate(
groups,
{
group_mean <- rnorm(1, mean = mu_alpha, sd = sigma_alpha)
errors <- rnorm(group_size, sd = sigma_epsilon)
tibble(y = group_mean + errors)
},
simplify = FALSE
) %>%
bind_rows(.id = "group") %>%
mutate(group = as_factor(group))
}
check_conf_int <- function(dataset, mu_alpha) {
mod <- lmer(y ~ (1 | group), dataset)
conf_int <- confint(mod, "(Intercept)")
between(mu_alpha, conf_int[, "2.5 %"], conf_int[, "97.5 %"])
}
# Set simulation parameters -----------------------------------------------
groups <- 48
group_size <- 16
mu_alpha <- 0
sigma_alpha <- 1
sigma_epsilon <- 0.25
runs <- 10000
# Make datasets -----------------------------------------------------------
set.seed(1)
# Takes 5 minutes
datasets <- replicate(
runs,
make_dataset(groups, group_size, mu_alpha, sigma_alpha, sigma_epsilon),
simplify = FALSE
)
# Run simulation ---------------------------------------------------------
# Takes 30 minutes
results <- map_lgl(datasets, check_conf_int, mu_alpha = mu_alpha)
tibble(
runs = seq_len(runs),
coverage = cummean(results)
) %>%
ggplot(aes(runs, coverage)) +
geom_line() +
geom_hline(linetype = "dashed", yintercept = 0.95) +
coord_cartesian(ylim = c(0.93, 0.97))
Below is the plot produced by the last code chunk:
Below is the output of $\tt{sessionInfo()}$:
R version 4.0.0 (2020-04-24)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Catalina 10.15.6
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.5.0 stringr_1.4.0 dplyr_0.8.5 purrr_0.3.4 readr_1.3.1
[6] tidyr_1.0.3 tibble_3.0.1 ggplot2_3.3.0 tidyverse_1.3.0 lme4_1.1-23
[11] Matrix_1.2-18
loaded via a namespace (and not attached):
[1] statmod_1.4.34 tidyselect_1.1.0 xfun_0.13 splines_4.0.0
[5] haven_2.2.0 lattice_0.20-41 colorspace_1.4-1 vctrs_0.3.0
[9] generics_0.0.2 htmltools_0.4.0 yaml_2.2.1 rlang_0.4.6
[13] nloptr_1.2.2.1 pillar_1.4.4 withr_2.2.0 glue_1.4.0
[17] DBI_1.1.0 dbplyr_1.4.3 modelr_0.1.7 readxl_1.3.1
[21] lifecycle_0.2.0 cellranger_1.1.0 munsell_0.5.0 gtable_0.3.0
[25] rvest_0.3.5 evaluate_0.14 labeling_0.3 knitr_1.28
[29] fansi_0.4.1 broom_0.5.6 Rcpp_1.0.4.6 backports_1.1.6
[33] scales_1.1.1 jsonlite_1.6.1 farver_2.0.3 fs_1.4.1
[37] hms_0.5.3 packrat_0.5.0 digest_0.6.25 stringi_1.4.6
[41] grid_4.0.0 cli_2.0.2 tools_4.0.0 magrittr_1.5
[45] crayon_1.3.4 pkgconfig_2.0.3 MASS_7.3-51.6 ellipsis_0.3.0
[49] xml2_1.3.2 reprex_0.3.0 lubridate_1.7.8 assertthat_0.2.1
[53] minqa_1.2.4 rmarkdown_2.1 httr_1.4.1 rstudioapi_0.11
[57] R6_2.4.1 boot_1.3-25 nlme_3.1-147 compiler_4.0.0
Updates
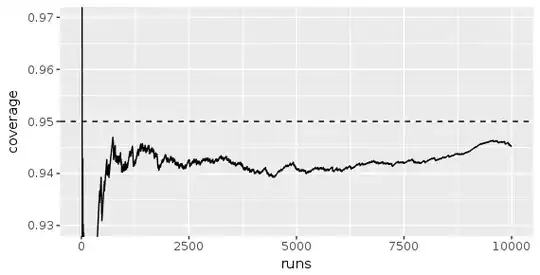
Below is the plot I get if I increase groups to 480:
This is the plot I get when I change group_size to 160: