I am a graduate student in animal science. I am comparing linear models that fit covariates of var1 and var2. These two covariates are decomposed from one quantity say F (inbreeding level of animal).
I decomposed F based on the cut-off number of generations in the pedigree used to define new inbreeding, e.g., 5, 10, 15 generations. For 5 generations, F = Fnew_5 + Fold_5, etc. My purpose is to just determine which cut-off threshold that is the best among candidates.
I want to compare model performance using different sets of var1 and var2.
y = mu + fixed_effects + b1var1 + b2var2 + e
where, y is body weight of animal, fixed_effects include sex, birth_year, age at measure. I chose to fit a fixed linear regression model due to its simplicity and my familiarity. I want to compare model performance (Aj.R2, RMSE, AIC, BIC) across all different ways to decompose the inbreeding F.
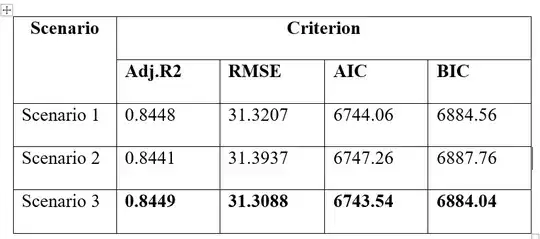
Results are:
In this case, scenario 3 showed slightly better than scenario 1 and 2 across all criteria.
My question is that does the model ranking change if I add another random effect (like animals) in the model? This random effect will be the same for all different models.
In other word, If I want just to see what is the best cut-off threshold to decompose inbreeding (F) in to new and old F, is fixed effect model sufficient to do so?
Thank you.