Your distribution $p_k \sim k^{-\alpha-1}$ for $k \geq k_{\text{min}}$, $k_{\text{min}} > 0$ is a truncated zeta distribution.
The distribution has no finite variance for $\alpha<2$ and the scaled sum will not approach a normal distribution.
However, you can apply a generalization of the central limit theorem. The limiting distribution of the following sum
$$S_n = \frac{ \sum_{i=1}^n (X_i-\mu_{X})}{n^{\frac{1}{\alpha}}} $$
will be a distribution of the stable distribution family with $\alpha = 1.2$.
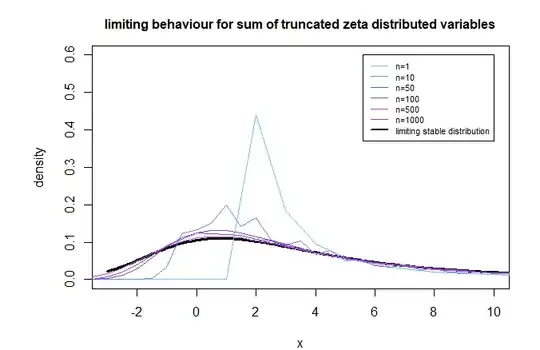
When we simulate this then it appears like the sum $S_n$ is approaching a stable distribution with $\beta = 1$ and $\gamma = 1$.
I guess (intuitively) that you can derive these $\beta$ and $\gamma$ by looking at the tails of the distribution whose asymptotic behavior is $$f(x) \approx \begin{cases} \frac{a}{\vert x \vert^{1+\alpha}} \quad \text{for} \quad x \to \infty \\
\frac{b}{\vert x \vert^{1+\alpha}} \quad \text{for} \quad x \to -\infty \end{cases} $$
where the $a$ and $b$ are constants depending on $\alpha$, $\beta$, $\gamma$ and $\delta$.
We can argue that $\beta = 1$ such that the weight in left tail will be zero ($b=0$).
We may probably argue something similar such that we get $\gamma = 1$ for non truncated distribution and $\gamma = 1/(1-P(X_{\text{truncated}} \leq k_{min}))^{1/\alpha}$ for the truncated distribution. But it is a bit based on intuition and handwavy. I have no good method for this yet to proof it with more rigor, but the computational result below shows that it probably works.
image:
code:
library(VGAM)
library(truncdist)
library(rmutil)
library(stabledist)
### alternative rzeta function because VGAM's qzeta and rzeta is slow
### here we create a table based on dzeta
ztable <- cumsum(VGAM::dzeta(1:10^7,1.2))
rzeta2 <- function(n,trunc = 0) {
u <- runif(n,c(0,ztable)[trunc+1],1)
u <- u[order(u)]
pos <- 1
x <- numeric()
for (i in 1:n) {
while(u[i]>ztable[pos]) {
pos = pos+1
}
x <- c(x,pos)
}
return(x)
}
### create a matrix with simulation results
ns <- 10^5
x <- matrix(rep(0,ns*6), ns)
y <- matrix(rep(0,ns*6), ns)
### simulate results with 6 different sample sizes
### non-truncated
set.seed(1)
for (i in 1:6) {
nsample <- c(1,10,50,100,500,1000)[i]
x[,i] <- replicate(ns, mean(rzeta2(nsample)))
}
### simulate results with 6 different sample sizes
### truncated
set.seed(1)
for (i in 1:6) {
nsample <- c(1,10,50,100,500,1000)[i]
y[,i] <- replicate(ns, mean(rzeta2(nsample,trunc = 1)))
}
### mean of non-truncated distribution
muzipf <- VGAM::zeta(1.2)/VGAM::zeta(2.2)
### mean of truncated distribution
mutrunc <- (muzipf - 1/VGAM::zeta(2.2))/(1-1/VGAM::zeta(2.2))
### plot results
plot(-100,-100, xlim = c(-3,10), ylim = c(0,0.6),
xlab = "x", ylab = "density", log = "")
### limiting stable distribution
beta <- 1
gamma <- 1
xs <- seq(-3,20,0.1)
ds <- dstable(xs , alpha = 1.2,
beta = beta,
gamma = gamma,
delta = muzipf+beta*gamma*tan(pi/2*1.2))
lines(xs,ds,lty = 1, lwd = 3)
### itterate the different sample sizes
for (i in 1:6) {
nsample <- c(1,10,50,100,500,1000)[i]
sep <- c(1,0.5,0.5,0.5,0.5,0.5)[i]
### scaling the distribution
xstable <- muzipf+(x[,i]-muzipf)*(nsample)^(1-1/1.2)
xstable <- xstable[(xstable>=-5)&(xstable<=15)]
### compute histogram
h <- hist(xstable, breaks = seq(-6,16,sep)-sep/2, plot = FALSE)
### plot histogram as curve
lines(h$mids,h$counts/ns/sep, col = hsv(0.5+i/16,0.5+i/16,1))
}
i <- c(1:6)
legend(10,0.6, c("n=1","n=10","n=50","n=100","n=500","n=1000","limiting stable distribution"),
lty = 1, col = c(hsv(0.5+i/16,0.5+i/16,1),"black"), lwd = c(rep(1,6),2),
xjust = 1 , cex = 0.7)
title("limiting behaviour for sum of zeta distributed variables")
### plot results
plot(-100,-100, xlim = c(-3,10), ylim = c(0,0.6),
xlab = "x", ylab = "density", log = "")
### limiting stable distribution
beta <- 1
gamma <- (1-dzeta(1,1.2))^(-1/1.2) # we increase gamma because the tail will be heavier
xs <- seq(-3,20,0.1)
ds <- dstable(xs , alpha = 1.2,
beta = beta,
gamma = gamma,
delta = mutrunc+beta*gamma*tan(pi/2*1.2))
lines(xs,ds,lty = 1, lwd = 3)
### itterate the different sample sizes
for (i in 1:3) {
nsample <- c(1,10,50,100,500,1000)[i]
sep <- c(1,0.5,0.5,0.5,0.5,0.5)[i]
### scaling the distribution
xstable <- mutrunc+(y[,i]-mutrunc)*(nsample)^(1-1/1.2)
xstable <- xstable[(xstable>=-5)&(xstable<=15)]
### compute histogram
h <- hist(xstable, breaks = seq(-6,16,sep)-sep/2, plot = FALSE)
### plot histogram as curve
lines(h$mids,h$counts/ns/sep, col = hsv(0.5+i/16,0.5+i/16,1))
}
i <- c(1:6)
legend(10,0.6, c("n=1","n=10","n=50","n=100","n=500","n=1000","limiting stable distribution"),
lty = 1, col = c(hsv(0.5+i/16,0.5+i/16,1),"black"), lwd = c(rep(1,6),2),
xjust = 1 , cex = 0.7)
title("limiting behaviour for sum of truncated zeta distributed variables")
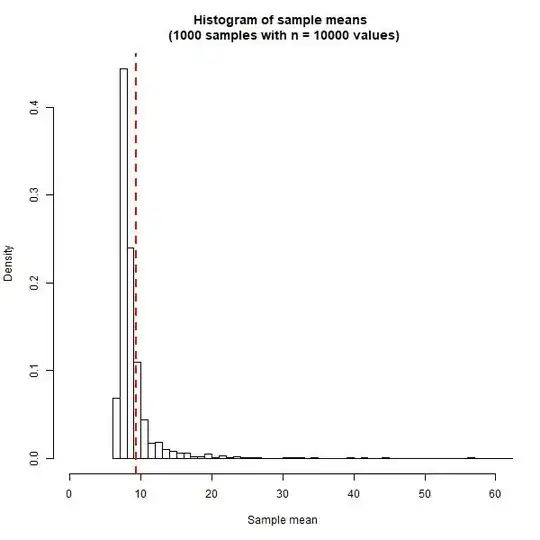
Thus, when I generate my samples, and take sample averages, I expect that these averages should be close to 9.36. However, when I plot the sampling distribution for the mean (i.e. the distribution of these sample averages), I get highly skewed distribution as shown below (total 1000 samples were generated):
Yes, as explained and shown above, the sample mean does not approach a normal distribution but instead an $\alpha$-stable distribution (which will be highly skewed and fat tailed)
But my question is, if I want to say that my results correspond to mean value 9.36 would that be right...
The results of the experimental sample distribution should correspond to the theoretical sample distribution. But the observed mean may indeed vary a bit from the theoretical mean.
...can I generate the samples so that the distribution of the sample averages would be symmetric around the theoretical mean?
You should not do that. The distribution of the sample averages is not symmetric. You can choose maybe a different population to sample from, but I can you have some reason to use the powerlaw.