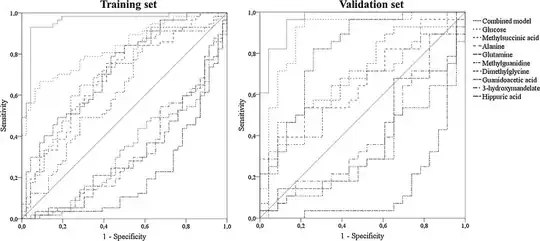
I have come across papers where they have calculated the AUROC for both a training and a testing set;
When I am using the package MLeval, I have used my training data set here
randomforestfit1 <- train(T2DS ~ .,
data = mod_train.newy,
method = "rf",
trControl = trainControl(method = "repeatedcv",
number = 10,
repeats = 5,
savePredictions= TRUE,
classProbs= TRUE,
verboseIter = TRUE))
##
x <- evalm(randomforestfit)
## get roc curve plotted in ggplot2
x$roc
## get AUC and other metrics
x$stdres
My AUROC for metabolites+ visceral fat + crp-1 is 0.82
My AUROC for visceral fat and crp-1 is 0.69
When using my validation set it is 0.88 and 0.86 respectively. I thought it was better to mention only the validation set rather than both. Please can anybody advise?