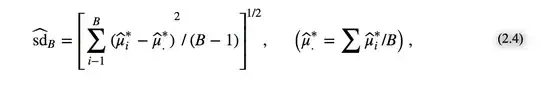I am having some difficulties reconciling a test for the difference in means using a standard test and the bootstrap. It would be great if someone could tell me what is going wrong in the code.
I run two regressions on two different samples and obtain the following:
$b_1 = 0.02 \hspace{3mm} (0.09)$
$b_2 = -0.03 \hspace{3mm} (0.08)$
There are 8500 observations in each sample.
Based on the answer here, the t-statistic can be calculated using the standard error $s$ below:
\begin{equation} s =\sqrt{\frac{(n_1 - p)s_1^2 + (n_2 - p)s_2^2}{n_1 + n_2 - 2p}} \end{equation} With one regressor $p$, I did this by hand and I find that $s = 0.09$, so that the t-statistic would be $0.55$.
However, I also try to bootstrap the standard errors, I get a much smaller standard error ($0.03$). To give an idea of the process I am using:
- Draw a bootstrap sample from group 1
- Estimate the model using the data in step 1
- Store the coefficient estimate as $b_1$
- Draw a bootstrap sample from group 2
- Estimate the model using the data in step 4
- Store the coefficient estimate as $b_2$
- Repeat the above 100 times
Then I perform a t-test on the two vectors containing $b_1$ and $b_2$. I suppose something is wrong in the steps above, but I don't know what it is. I am using bootstrap because I cannot include interaction terms in my model.
Thank you.
EDIT Here is the code I am using.
B<-50
t.vector1<-vector(length=B)
t.vector2<-vector(length=B)
for (j in 1:B){
boot.dat1<-data1[sample(nrow(data1),nrow(data1),replace=TRUE),]
boot.mod1<-rdrobust(y=boot.dat1$y,x=boot.dat1$x)
boot.dat2<-data2[sample(nrow(data2),nrow(data2),replace=TRUE),]
boot.mod2<-rdrobust(y=boot.dat2$y,x=boot.dat2$x,h=(boot.mod1$bws[1,1])) # local linear regression, so I use the same bandwidth as mod1
t.vector1[j]<-boot.mod1$Estimate[1,1]
t.vector2[j]<-boot.mod2$Estimate[1,1]
}
ttest<-t.test(t.vector1,t.vector2)
ttest$stderr
[1] 0.02295152