This might be a bit long so I will reply in another answer.
Following the comment to my previous answer I will attempt a solution to the following problem: fit an additive model with a shared smooth trend effect subject to boundary constraint and a random Id intercept.
Extra penalty in matrix augmentation form
In the comments above I mentioned that strategy 1 of my previous answer can be used to achieve the constrained fit in a GAMM setting. This becomes clear if we write the extra penalty solution in augmented matrix form (in what follows I will use the same notation as in my previous answer). We can say that:
$$
\min_{c} S_{p} = \|W^{1/2} (y_{p} - B_{p}c)\|^{2} + \lambda \|D_{d} c\|^{2}
$$
where $c$ is a $(m \times 1)$ vector of unknown spline coefficients, $y_{p}$ is a $((n+ 2) \times 1)$ vector obtained stacking the observed $y$ and boundary values $v(x_{0})$, $B_{p}$ is a $((n+2) \times m)$ B-splines basis matrix obtained placing matrices $B$ and $\Gamma$ on top of each other, $W^{1/2}$ is a $((m + 2) \times (m+2))$ diagonal matrix with first $m$ non-zero elements equal to 1 and last two equal to $\sqrt{\kappa}$ and $D_{d}$ is a $d$ order finite difference matrix operator (the penalty matrix is equal to $P = D_{d}^{\top} D_{d}$).
P-splines as mixed models
P-splines (and all the penalized smoothing techniques included in the s() function of the mgcv package) can be written in a linear 'mixed model form'. For P-splines different re-parameterization are possible (see e.g. par 10 of Eilers et al. (2015)). We can for example define
$$
\begin{array}{ll}
X = [1, x_{p}^{1}, ..., x_{p}^{d-1}] \\
Z = B_{p}D_{d}^{\top} (D_{d}D_{d}^{\top})^{-1}
\end{array}
$$
where $x_{p}$ is the $(m+2)$ vector of time points including the last two boundary ordinates. With this in mind we can then rewrite the normal equations for the min problem above as follows
(see also this):
$$
\left[
\begin{array}{lll}
X^{\top} W X & X^{\top} W Z \\
Z^{\top} W X & Z^{\top} W Z + \lambda I
\end{array}
\right]
\left[
\begin{array}{ll}
\beta\\ b
\end{array}
\right]
=
\left[
\begin{array}{ll}
X^{\top} W y_{p} \\ Z^{\top} W y_{p}
\end{array}
\right]
$$
where $\lambda$ is still the smoothing parameter and it is equal to the ratio of variances $\sigma^{2}/\tau^{2}$ with $b \sim N(0, \tau^{2} I)$ and $\epsilon \sim N(0, \sigma^{2} I)$.
Include random intercept
To solve the original problem, we would like to include also a random intercept. This can be achieved by modifying the form of the Z matrix as follows (see also this link ):
$$
Z = \left(
\begin{array}{lll}
Z_{1} ,& \texttt{1}_{1},& 0,& \dots ,& 0 \\
Z_{2} ,& 0,& \texttt{1}_{2},& \dots ,& 0 \\
\vdots ,& \vdots,& \vdots,& \vdots,& \vdots \\
Z_{J} ,& \dots,& \dots, & \dots,& \texttt{1}_{J}
\end{array}
\right)
$$
where
$\texttt{1}_{j}$ is a $((n_{j} + 2) \times 1)$ vector of ones used to model the $j-$th subject-specific intercept. Of course this also 'adds' $J$ elements to the vector of random effects $b$ with $\text{Cov}(b) = \begin{pmatrix}
\tau^2 \boldsymbol{I} & 0 \\
0 & \sigma_{\texttt{1}}^2 \boldsymbol{I}
\end{pmatrix}$
Small R-code
I will suppose here that you have a function for the definition of the B-spline matrices and their mixed model representation. I left comments and references in the code. In principle, I think this can be achieved within the mgcv package but unfortunately I do not know the package well enough. Instead, I will use nlme package (on which I thing mgcv is written on at least partially).
#####################
# Utility functions #
#####################
Conf_Bands = function(X, Z, f_hat, s2, s2.alpha, alpha = 0.975)
{
# cit: #http://halweb.uc3m.es/esp/Personal/personas/durban/esp/web/cursos/Maringa/gam-markdown/Gams.html#26_penalized_splines_as_mixed_models
C = cbind(X, Z)
lambda = s2/s2.alpha
D = diag(c(rep(0, ncol(X)), rep(lambda, ncol(Z))))
S = s2 * rowSums(C %*% solve(t(C) %*% C + D) * C)
CB_lower = f_hat - qnorm(alpha) * sqrt(S)
CB_upper = f_hat + qnorm(alpha) * sqrt(S)
CB = cbind(CB_lower, CB_upper)
CB
}
basesMM = function(B, D, dd, ns, x)
{
# NB: needs to be modified if n_{j} is different for some j
Z0 = B %*% t(D) %*% solve(D %*% t(D))
X0 = outer(x, 1:(dd-1), '^')
Z = do.call('rbind', lapply(1:ns, function(i) Z0))
X = do.call('rbind', lapply(1:ns, function(i) X0))
return(list(X = X, Z = Z))
}
#########################
# Utility functions end #
#########################
# Simulate some data
set.seed(2020)
xmin = 1
xmax = 9
m = 100
x = seq(xmin, xmax, length = m)
ys = sin((x^2)/10)
ns = 3
y = ys + rnorm(m) * 0.2
yl = c(-2 + y, -0 + y, 2 + y)
sb = factor(rep(1:3, each = m))
dat = data.frame(y = yl, x = rep(x, ns), sub = sb)
# Boundary conditions
bx = c(0, 10)
by = c(-0, -0)
xfine = seq(bx[1], bx[2], len = m * 2)
# Create bases
# see https://onlinelibrary.wiley.com/doi/10.1002/wics.125
bdeg = 3
nseg = 25
dx = (bx[2] - bx[1]) /nseg
knots = seq(bx[1] - bdeg * dx, bx[2] + bdeg * dx, by = dx)
B0 = bbase(x, bx[1], bx[2], nseg, bdeg)
nb = ncol(B0)
Gi = bbase(bx, bx[1], bx[2], nseg, bdeg)
Bf = bbase(xfine, bx[1], bx[2], nseg, bdeg)
# Penalty stuffs
dd = 3
D = diff(diag(1, nb), diff = dd)
kap = 1e8
# Augmented matrix
Bp = rbind(B0, Gi)
# Mixed model representation for lme
# see https://www.researchgate.net/publication/290086196_Twenty_years_of_P-splines
yp = do.call('c', lapply(split(dat, dat$sub), function (x) c(x$y, by)))
datMM = data.frame(y = yp)
mmBases = basesMM(Bp, D, dd, ns, x = c(x, bx))
datMM$X = mmBases$X
datMM$Z = mmBases$Z
datMM$w = c(rep(1, m), 1/kap, 1/kap)
datMM$Id = factor(rep(1, ns * (m + 2)))
datMM$sb = factor(rep(1:ns, each = m + 2))
# lme fit:
# https://www.researchgate.net/publication/8159699_Simple_fitting_of_subject-specific_curves_for_longitudinal_data
# https://stat.ethz.ch/pipermail/r-help/2006-January/087023.html
# https://stats.stackexchange.com/questions/30970/understanding-the-linear-mixed-effects-model-equation-and-fitting-a-random-effec
fit = lme(y ~ X, random = list(Id = pdIdent(~ Z - 1), sb = pdIdent( ~ w - 1)), data = datMM, weights = ~w)
# Variance components
s2 = fit$sigm ^ 2
s2.alpha = s2 * exp(2 * unlist(fit$modelStruct)[1])
# Extract coefficients + get fit + value at boundaries
X0 = datMM$X[1:(m+2), ]
Z0 = datMM$Z[1:(m+2), ]
beta.hat = fit$coef$fixed
b.hat = fit$coef$random
f.hat = cbind(1, X0[1:m, ]) %*% beta.hat + Z0[1:m, ] %*% t(b.hat$Id)
f.hatfine = cbind(1,basesMM(Bf, D, dd, ns = 1, x = xfine)$X) %*% beta.hat + basesMM(Bf, D, dd, ns = 1, x = xfine)$Z %*% t(b.hat$Id)
f.cnt = cbind(1, X0[-c(1:m), ]) %*% beta.hat + Z0[-c(1:m), ] %*% t(b.hat$Id)
fit_bands = Conf_Bands(cbind(1, X0[1:m, ]) , Z0[1:m, ], f.hat, s2, s2.alpha)
# Plots fits
par(mfrow = c(2, 1), mar = rep(2, 4))
plot(rep(x,ns), yl, xlim = range(c(x, bx) + c(-0.5, 0.5)), main = 'Fitted curves', col = as.numeric(dat$sub), pch = 16)
abline(h = 0, lty = 3)
lines(x, f.hat[1:m] + fit$coefficients$random$sb[1], col = 8, lwd = 2)
lines(x, f.hat[1:m] + fit$coefficients$random$sb[2], col = 8, lwd = 2)
lines(x, f.hat[1:m] + fit$coefficients$random$sb[3], col = 8, lwd = 2)
# Plot smooths
plot(x, f.hat, type = 'l', main = 'Smooth-term', xlim = range(c(x, bx) + c(-0.5, 0.5)), ylim = range(fit_bands + c(-0.5, 0.5)))
rug(knots[knots <= bx[2] & knots >= bx[1]])
polygon(x = c(x, rev(x)), y = c(fit_bands[, 1], rev(fit_bands[, 2])), lty = 0, col = scales::alpha('black', alpha = 0.25))
abline(h = by)
points(bx, f.cnt, pch = 16)
lines(xfine, f.hatfine, col = 2, lty = 2)
legend('topleft', legend = c('Smooth', 'Extrapolation', 'Constraint'), col = c(1, 2, 1), lty = c(1, 2, 0), pch = c(-1, -1, 16))
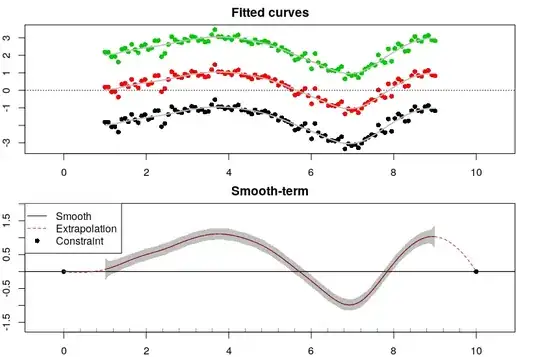
I hope everything here is correct (if you find some mistake, things that are not clear or suggestions please let me know). Finally, I hope my answer will be useful.