It seems that the OP is mainly concerned with conducting a t-test for unequal sample sizes.
However, imbalanced sample sizes are in general not a huge problem when applying the t-Test. The t-test is actually very stable against sample size characteristics as deviations from normal distribution and imbalanced sample sizes (if sample sizes are large enough!). Small simulations are following to illustrate this:
p.value <- c()
n.sim <- 1e5
for(i in 1:n.sim){
x1 <- rnorm(n=100,mean=1,sd=1)
x2 <- rnorm(n=1000,mean=1,sd=1)
p.value[i] <- t.test(x1, x2)$p.value
}
sum(p.value<0.05)/n.sim
> [1] 0.05083
As you see, no increase of type I error rate. I have assumed that both samples come from a normal distributions, though. Let's consider a completely different distribution, e.g. two very different Gamma distributions but with the same mean (mean is shape divided by rate):
p.value <- c()
n.sim <- 1e5
for(i in 1:n.sim){
x1 <- rgamma(n=100,shape=1,rate=1)
x2 <- rgamma(n=1000,shape=10,rate=10)
p.value[i] <- t.test(x1, x2)$p.value
}
sum(p.value<0.05)/n.sim
> [1] 0.05749
Slightly increased type I error rate. Whether this is serious enough to prevent from using a t-test is debatable. Taking a look at the distribution of p-values might be insightful, too. Under the null hypothesis, p-values should be uniformly distributed, and it looks pretty much like it:
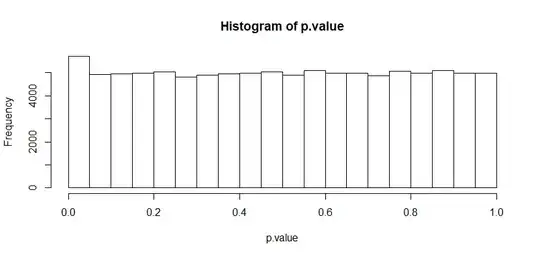
Note that this problem will become more serious, when your sample sizes are smaller than what I assumed, e.g. for the last example with the sample sizes being 10 and 100 instead, the type I error rate becomes 0.10177.
What happens if we take subsamples from the larger group, calculate the p.value each time, and then average these p.values?
p.value <- c()
pvalue2 <- c()
n.sim <- 1e5
for(i in 1:n.sim){
x1 <- rgamma(n=100,shape=1,rate=1)
x2 <- rgamma(n=1000,shape=10,rate=10)
for(j in 1:10){
x2.sub <- x2[((j-1)*100+1):(j*100)]
pvalue2[j] <- t.test(x1, x2.sub)$p.value
}
p.value[i] <- mean(pvalue2)
}
sum(p.value<0.05)/n.sim
> [1] 0.03909
Seems something might be off with the type I error rate. But it gets really interesting if we take a look at the distribution of p-values. 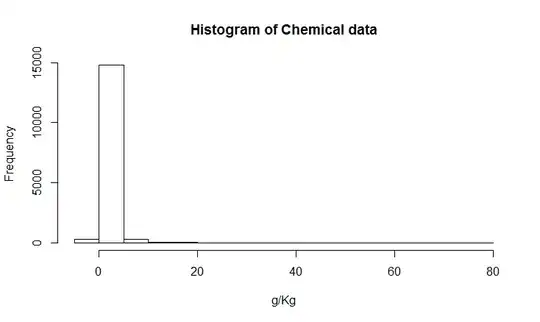 Doesn't look like a uniform distribution at all anymore! With imbalanced sample sizes we are far better off than with taking the mean of p-values for several subsamples.
Doesn't look like a uniform distribution at all anymore! With imbalanced sample sizes we are far better off than with taking the mean of p-values for several subsamples.

