In Edward Frees' book Predictive Modeling Applications in Actuarial Science, Volume 2 the first chapter goes over how to build a frequency GLM model (using a Poisson distribution) on sample auto-insurance data.
To test the data, they used cross validation to find a best fit model. I decided to check a QQ-plot to see if the deviance residuals followed a normal distribution.
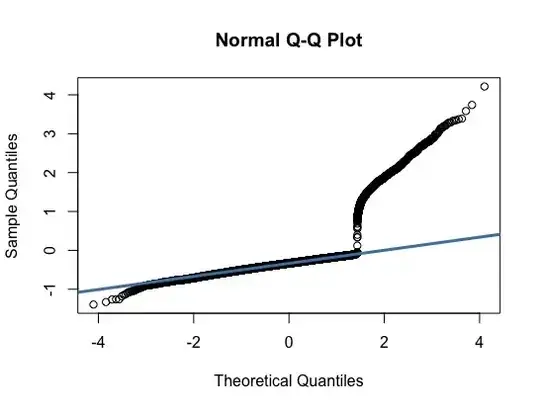
According to a QQ-plot of the deviance residuals, the left and right tails are skewed and don't follow the typical 45-degree line in a normal distribution. (See QQ-plot picture for further reference). Under the assumption that the cross-validation results were satisfactory, does that imply a good distribution fit (since they're both also testing for goodness-of-fit)?
The modeling equation is: fq.m <- glm(clm.count ~ year + ncd.level + drv.age.gr2 + yrs.lic + region.g1 + prior.claims, family = poisson(link = "log"), data = dta, subset = train, offset = log(exposure)).
And the code is below:
Call:
glm(formula = clm.count ~ year + ncd.level + drv.age.gr2 + yrs.lic +
region.g1 + prior.claims, family = poisson(link = "log"),
data = dta, subset = train, offset = log(exposure))
Deviance Residuals:
Min 1Q Median 3Q
-1.3934 -0.4462 -0.3320 -0.2188
Max
4.2149
Coefficients:
Estimate Std. Error
(Intercept) -1.21777 0.08411
year2010 -0.55021 0.07352
year2011 -0.65080 0.06707
year2012 -0.08010 0.05361
ncd.level2 -0.46911 0.20658
ncd.level3 -0.10167 0.06387
ncd.level4 -0.28405 0.08414
ncd.level5 -0.39079 0.09790
ncd.level6 -0.58293 0.09247
drv.age.gr218-22 0.53856 0.27462
drv.age.gr223-27 0.49162 0.11650
drv.age.gr228-32 0.29560 0.08632
drv.age.gr233-37 0.14536 0.07988
drv.age.gr243-47 0.17591 0.08158
drv.age.gr248-52 0.18371 0.08391
drv.age.gr253-57 0.17848 0.09278
drv.age.gr258-62 0.10453 0.10905
drv.age.gr263+ -0.00787 0.12571
yrs.lic2 -0.20783 0.06866
yrs.lic3 -0.36337 0.08091
yrs.lic4 -0.31022 0.08785
yrs.lic5 -0.47068 0.10506
yrs.lic6 -0.74737 0.13597
yrs.lic7 -0.54567 0.16332
yrs.lic8+ -0.48231 0.23087
region.g1R1 -0.55144 0.12420
region.g1R2 -0.40082 0.13897
region.g1R3 -0.32314 0.06592
region.g1R4 -0.25546 0.08384
region.g1R5 -0.18492 0.08693
region.g1R6 -0.08658 0.06876
region.g1R7 -1.05186 0.19124
region.g1R8 0.11225 0.09422
prior.claims 0.13521 0.01432
z value Pr(>|z|)
(Intercept) -14.477 < 2e-16 ***
year2010 -7.484 7.22e-14 ***
year2011 -9.704 < 2e-16 ***
year2012 -1.494 0.135175
ncd.level2 -2.271 0.023159 *
ncd.level3 -1.592 0.111418
ncd.level4 -3.376 0.000736 ***
ncd.level5 -3.992 6.56e-05 ***
ncd.level6 -6.304 2.90e-10 ***
drv.age.gr218-22 1.961 0.049866 *
drv.age.gr223-27 4.220 2.44e-05 ***
drv.age.gr228-32 3.425 0.000616 ***
drv.age.gr233-37 1.820 0.068820 .
drv.age.gr243-47 2.156 0.031069 *
drv.age.gr248-52 2.189 0.028573 *
drv.age.gr253-57 1.924 0.054387 .
drv.age.gr258-62 0.959 0.337791
drv.age.gr263+ -0.063 0.950080
yrs.lic2 -3.027 0.002470 **
yrs.lic3 -4.491 7.09e-06 ***
yrs.lic4 -3.531 0.000413 ***
yrs.lic5 -4.480 7.46e-06 ***
yrs.lic6 -5.497 3.87e-08 ***
yrs.lic7 -3.341 0.000834 ***
yrs.lic8+ -2.089 0.036697 *
region.g1R1 -4.440 9.00e-06 ***
region.g1R2 -2.884 0.003925 **
region.g1R3 -4.902 9.47e-07 ***
region.g1R4 -3.047 0.002310 **
region.g1R5 -2.127 0.033405 *
region.g1R6 -1.259 0.207957
region.g1R7 -5.500 3.79e-08 ***
region.g1R8 1.191 0.233482
prior.claims 9.439 < 2e-16 ***
---
Signif. codes:
0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’
0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 9847.2 on 24494 degrees of freedom
Residual deviance: 9282.3 on 24461 degrees of freedom
AIC: 13150
Number of Fisher Scoring iterations: 6