So i was just having a nice evening trying to simulate some hierarchical models by myself and estimate their parameters. First model I tried to simulate and estimate was $$y_{ij}=\beta_0+\varepsilon_{ij},$$ where $b_0=\gamma_{00}+u_{0j}$. So actually what I was estimating was $y_{ij}=\gamma_{00}+u_{0j}+\varepsilon_{ij}$. I have generated some data and estimated parameters $\gamma_{00}$ and variances of error terms and everything was fine:
set.seed(1)
N <- 100
nj <- 100
g00 <- 10
e <- rnorm(N*nj)
j <- c(sapply(1:N, function(x) rep(x, nj)))
uj <- c(sapply(1:N, function(x)rep(rnorm(1), nj)))
d <- data.frame(j, y=g00+uj+e)
library(nlme)
lme(y~1, data=d, random=~1|j)
Linear mixed-effects model fit by REML
Data: d
Log-restricted-likelihood: -14520.94
Fixed: y ~ 1
(Intercept)
10.00215
Random effects:
Formula: ~1 | j
(Intercept) Residual
StdDev: 0.7752422 1.012683
Number of Observations: 10000
Number of Groups: 100
Then I tried different model:
$$y_{ij}=\beta_0+\beta_1 x_{ij}+\varepsilon_{ij},$$
where $\beta_0=\gamma_{00}+u_{0j}$ and $\beta_1=\gamma_{10}+u_{1j}$. So I had to estimate the equation
$$y_{ij}=\gamma_{00}+u_{0j}+(\gamma_{10}+u_{1j}) \cdot x_{ij}+\varepsilon_{ij}=\gamma_{00}+\gamma_{10}x_ij+u_{0j}+u_{1j}x_{ij}+\varepsilon_{ij}.$$
I did the same thing that I had before, but this time lme did not converge. I tried this, but it didn't seem to work.
g10 <- 10
u0j <- uj
u1j <- c(sapply(1:N, function(x)rep(rnorm(1), nj)))
x1 <- rnorm(N*nj)
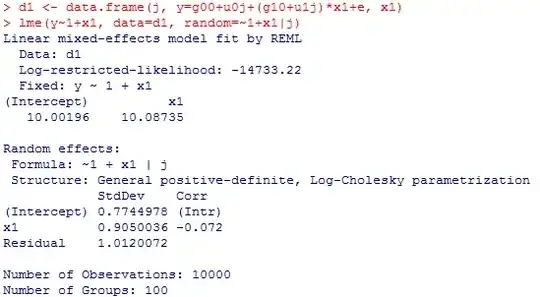
d1 <- data.frame(j, y=g00+u0j+(g10+u1j)*x1, x1)
lme(y~1+x1, data=d1, random=~1+x1|j)
Here is what last call to lme spit out:
Error in lme.formula(y ~ 1 + x1, data = d1, random = ~1 + x1 | j) :
nlminb problem, convergence error code = 1
message = false convergence (8)
What can you suggest me to do? Maybe the problem is with my model specification, redundant parameters, singular matrix or something else?