In order to be clear about what can be said, in the following analysis capital letters refer to random variables (not their realizations).
You have $N$ independent random variables $X_i$ with identical Normal$(0,1)$ distributions. From them you form $m$ new variables $Y_1=X_1+\cdots+X_n$ and so on. Very generally, you can form new variables out of old by choosing any coefficients $a_{ij}$ you like and set
$$Y_i = \sum_{k=1}^N a_{ik} X_k$$
for $i=1, 2, \ldots, m.$
The question asks about the variance of the "set" of $Y_i$ (its square root would be the standard deviation). I believe we have to understand that as referring to some multiple of the expression
$$\sum_{i=1}^m (Y_i - \bar Y)^2\tag{*}$$
where $\bar Y = (Y_1 + \cdots + Y_m)/m.$ (The usual multiples are $1/m$ and $1/(m-1).$ A constant multiple presents no problem, because ultimately it will just multiply the standard deviation.)
Let $\mathbb{A}=(a_{ik})$ be the $m\times N$ matrix of coefficients and let $\mathbb{P} = \mathbb{I}_m - \mathbf{1}^\prime \mathbf{1}$ be the matrix the removes the column means from $\mathbb A$ when multiplied (on the left). In these terms, $(*)$ is the quadratic form in the vector $\mathbf{X}=(X_1,X_2, \ldots, X_N)$ with matrix
$$\mathbb{Q} = \mathbb{A}^\prime \mathbb{P} \mathbb{A}.$$
Because $\mathbb Q$ is symmetric and positive-semidefinite, it can be diagonalized with an orthogonal transformation. This means we may express the form as
$$\sum_{i=1}^m (Y_i - \bar Y)^2\tag{*} = \sum_{j=1}^{N} \lambda_j Z_j^2$$
with all the $\lambda_j$ non-negative and, because (a) the $Z_j$ are linear combinations of the independent Normal variables $X_k$ and (b) these linear combinations are orthogonal, the $Z_j$ are independent standard Normal variables. Their squares $Z_j^2$ all have $\chi^2(1)$ distributions (by definition), exhibiting $(*)$ as a linear combination of independent $\chi^2(1)$ distributions. Its square root (multiplied by a suitable constant) gives the distribution of the sd of the $Y_i.$
Because a $\chi^2(1)$ distribution is a multiple of a $\Gamma(1/2)$ distribution, the analysis at https://stats.stackexchange.com/a/72486/919 shows there is no simple expression for it. Even in the case where $\mathbb A$ is given as in the question, all we can say generally is that $N-m$ of the $\lambda_j$ are nonzero and that their range increases as $N-m$ increases (precluding simple general approximations that assume the nonzero $\lambda_j$ are equal). It scarcely seems worthwhile to work out formulas for the $\lambda_j$ in terms of $N$ and $m,$ although surely that could be done.
The foregoing analysis does suggest the distribution might be approximated by some chi distribution (which governs the square root of a chi-squared variable). By matching moments of the chi-squared distributions (before taking square roots), the mean would be
$$\mu = \frac{1}{\sqrt{N-n}} \sum_i \lambda_i$$
and the variance would be
$$\sigma^2 = \frac{2}{N-n} \sum_i \lambda_i^2.$$
Therefore the approximating Gamma scale parameter would be
$$\theta = \frac{\mu}{\sigma^2}$$
and the shape parameter would be
$$k = \frac{\mu}{\theta} = \frac{\mu^2}{\sigma^2}.$$
The approximation is exact when $m=1$ and appears to work well for large $N$ and $n$ not close to $N/2.$ Here is an example for $N=39$ and $m=10:$
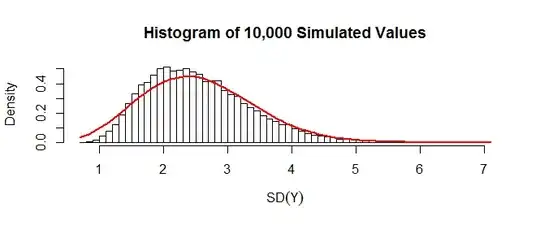
The red curve plots the density of the $\chi(2.200655, 1/17.80457)$ distribution (the parameters are shape and scale, respectively). Qualitatively this approximation is ok. When $N$ is five times larger (keeping $m=10$ as before), the approximation (now with a $\chi(13.70688, 1/9.449866)$ distribution) is excellent:
This R code creates a matrix $A:$
N <- 39*5
n <- 10
A <- outer(seq_len(N-n+1), seq_len(N), function(i,j) ifelse(i <= j & j < i+n, 1, 0))
A simulation of the standard deviations then is straightforward:
n.sim <- 1e4
X <- matrix(rnorm(n.sim * N), N)
Y <- A %*% X
S2 <- apply(Y, 2, sd)
Finding the eigenvalues $\lambda_i$ requires forming the matrix $\mathbb Q:$
m <- dim(A)[1]
P <- diag(rep(1,m)) - matrix(1/m, m, m)
Q <- t(A) %*% P %*% A
lambda <- zapsmall(eigen(Q, only.values=TRUE)$values)
lambda <- lambda[lambda > 0]
The last figure was constructed from the simulated SDs stored in S2 and eigenvalues stored in lambda:
mu <- sum(lambda) / sqrt(N-n)
sigma2 <- 2 * sum(lambda^2) / (N-n)
theta <- sigma2/mu
k <- mu/theta
hist(S2, freq=FALSE, breaks=50, xlab=expression(SD(Y)),
main="Histogram of 10,000 Simulated Values")
curve(2*x*sqrt(N-n)*dgamma(sqrt(N-n)*x^2, k, 1/theta), add=TRUE, col="Red", lwd=2)

