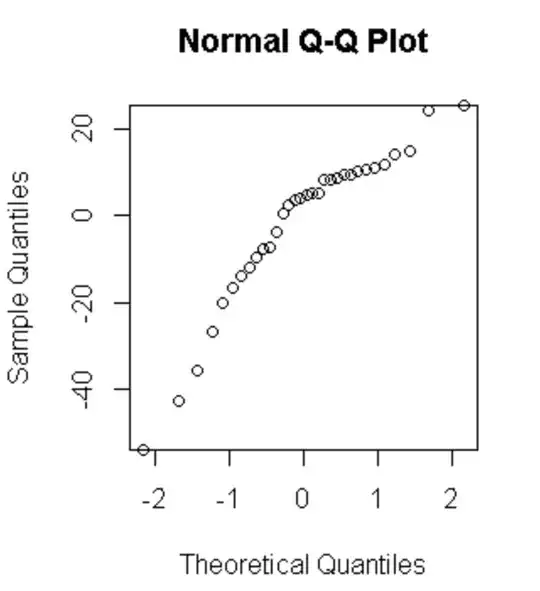
I am currently running a Shapiro-Wilk test on the residuals of a repeated ANOVA model. The repeated-measures ANOVA was conducted to measure two main effects A & B. The ANOVA summary is listed below.
Error: X
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 31 127893 4126
Error: X:A
Df Sum Sq Mean Sq F value Pr(>F)
A 1 2435 2436 7.122 0.012 *
Residuals 31 10601 342
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: X:B
Df Sum Sq Mean Sq F value Pr(>F)
B 1 362 361.6 1.915 0.176
Residuals 31 5854 188.8
Error: X:A:B
Df Sum Sq Mean Sq F value Pr(>F)
A:B 1 548 548.3 4.164 A:B 0.0499 *
Residuals 31 4081 131.7
Residuals
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 64 16940 264.7
The leve's test result indicated F(3,188) = 1.0945, p = 0.3527) that there is no significant evidence to suggest that variance across groups is different.
The Shapiro-Wilk test results are
data: aov_residuals_A
W = 0.89639, p-value = 0.005018
data: aov_residuals_B
W = 0.97361, p-value = 0.6045
data: aov_residuals_XY
W = 0.96834, p-value = 0.4549
Shapiro-Wilk test indicates that residuals of A are not normally distributed. But when I plot the qqnorm for the residuals, it looks similar. Is it ok if I report that the data is normally distributed based on the qqnorm or should I perform some other test other than ANOVA.