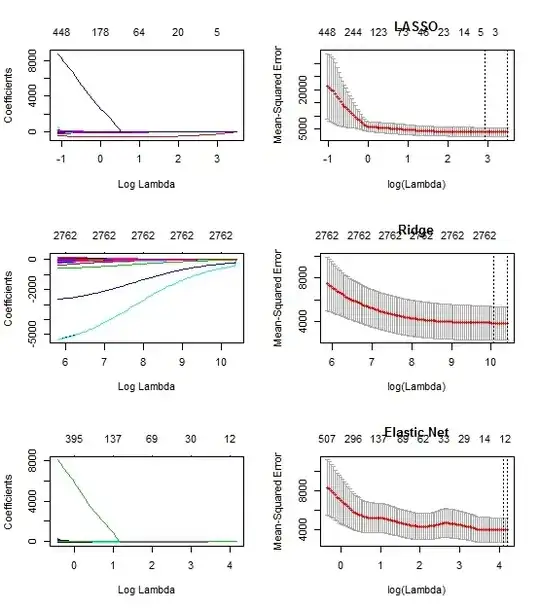
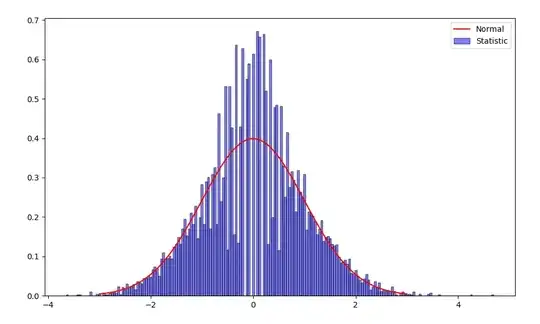
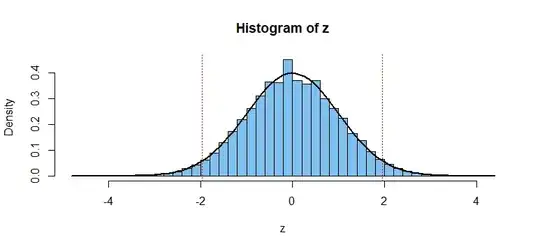
I saw in a few places (e.g. here) when you compare proportions of 2 samples, under a null hypothesis that they are equal, you eventually get to this:
$$ \frac{\bar X - \bar Y}{\sqrt{P(1-P)(\frac{1}{n} + \frac{1}{m})}} \sim N(0,1) $$
At which point there's a mental "jump" where you estimate $P$ from the total of two samples, and stick it in the above formula, i.e.:
$$ \hat P = \frac{\sum x_i + \sum y_i}{n + m} \\ \frac{\bar X - \bar Y}{\sqrt{\hat P(1- \hat P)(\frac{1}{n} + \frac{1}{m})}} \sim N(0,1) $$
My question is why is it legal to simply stick $\hat P$ instead of $P$ and still assume that it distributes on the standard normal distribution. Is there any proof of this?
UPDATE:
So I tried to simulate it myself, and indeed, when $n \neq m$, the histogram of the proportion statistic looks to fit very well the standard normal distribution.
However, if $n = m$, there seems to be a gap opening in the middle of the distribution:
Code (in Python):
import numpy as np
from scipy import sqrt, stats
from matplotlib import pyplot as plt
# Statistic
p = 0.2
n = 700
m = 300
X = np.random.binomial(n, p, 10000)
Y = np.random.binomial(m, p, 10000)
x_bar = (1/n) * X
y_bar = (1/m) * Y
est_p = (1/(n+m)) * (X + Y)
var = est_p * (1 - est_p) * (1/n + 1/m)
statistic = (x_bar - y_bar)/(sqrt(var))
plt.hist(statistic, density=1, color='blue', edgecolor='black', bins=200, alpha=0.5, label='Statistic')
# Normal
mu = 0
variance = 1
sigma = sqrt(variance)
x = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
plt.plot(x, stats.norm.pdf(x, mu, sigma), color='red', label='Normal')
plt.legend(loc='upper right')
plt.show()