I am trying to transform my GARCH standardized residuals to PITs in order to use them in a copula. The following code has been so far applied:
gjr_garch = arch_model(log_r['DAX'].dropna(), mean='AR', lags=8, vol='garch', p=1, o=1, q=1, dist='t')
dax_test = gjr_garch.fit()
dax_test.summary()
dax_dof = dax_test.tvalues['nu']
dax_sigma = dax_test.conditional_volatility
dax_innovations = dax_test.resid / dax_test.conditional_volatility
dax_innovations_std = (dax_innovations - np.mean(dax_innovations)) / np.std(dax_innovations)
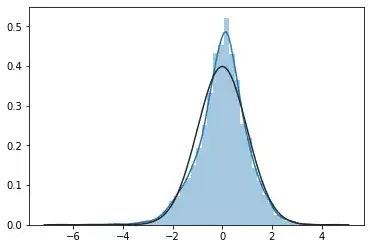
sns.distplot(dax_innovations_std.dropna(), hist=True,fit=stats.norm)
Next, I just take the CDF of the standardized innovations with their respective d.o.f. estimated by the GARCH model.
dax_uni = t.cdf(dax_innovations_std, dax_dof)
I convert them to pd.Series just so I can dropna()
dax_uni = pd.Series(data=dax_uni)
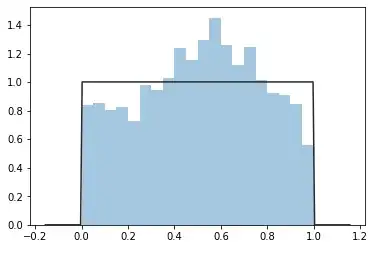
sns.distplot(dax_uni.dropna(), bins=20, fit=stats.uniform, kde=False)
In fact, I've done this for not only GJR-GARCH but also for AR-TARCH AR-EGARCH models with AR means ranging from 0 to 10. None of the Probability Integral Transforms pass the Kolmogorov-Smirnov test for Unif(0,1).
Any ideas why can't I get uniformly distributed probability transforms?