I'm trying to replicate the following code from SAS in R:
proc genmod data=skinny ;
class personid ;
model sample1_totalSpermCount_1 = samplePerson_1_byr
deepgen1935c totalchildren_1935 / dist=normal link = identity;
repeated subject=personid / type=exch;
where deepgen_1935>=3;
run;
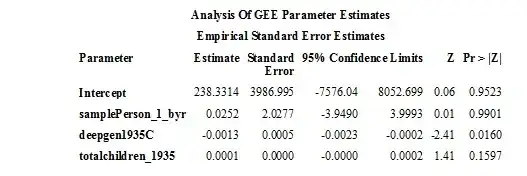
Here is the results in SAS:
Here is my code in R:
skinny.gee3 <- gee(sample1_totalSpermCount_1 ~ samplePerson_1_byr + deepgen1935.c + totalchildren_1935,
id = PersonID,
data = clean[clean$DeepGen_1935 >= 3, ],
family = gaussian,
corstr = "exchangeable")
I received an error, saying:
Warning message: In gee(sample1_totalSpermCount_1 ~ samplePerson_1_byr + deepgen1935.c + : Working correlation estimate not positive definite
And here is the results in R:
Coefficients:
Estimate Naive S.E. Naive z Robust S.E. Robust z
(Intercept) 1376.0792841 1659.6117221 0.8291574 1906.1089688 0.7219311
samplePerson_1_byr -0.5671856 0.8434850 -0.6724312 0.9694925 -0.5850335
deepgen1935.c -12.4639418 7.0401018 -1.7704207 4.3967397 -2.8348146
totalchildren_1935 0.6572002 0.6458183 1.0176240 0.4139229 1.5877359
I can see that the working correlation in SAS is different than in R. Is there an explanation why SAS's GEE run and R's GEE did not?
Thank you!