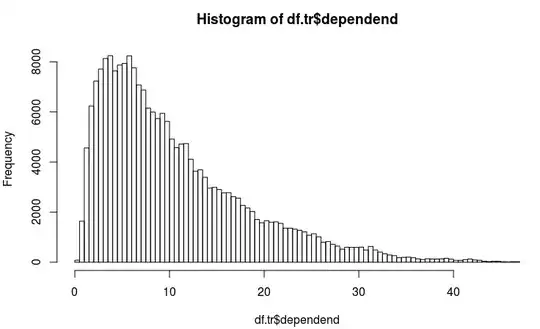
So we have a regression challenge here in which the dependent variable looks like this:
It is overdispersed as mean = 10 and variance = 60 so we think quasi-poisson is best?
We use the glm() functions in R.
We tried all of the below alternatives
m1g <- glm(round(dependend) ~ x1 * x2, data = df.tr )
m1g1 <- glm(dependend ~ x1 * x2, data = df.tr )
m1p <- glm(round(dependend) ~ x1 * x2, data = df.tr, family = "poisson" )
m1qp <- glm(round(dependend) ~ x1 * x2, data = df.tr, family = "quasipoisson" )
m1l <- glm(log(dependend) ~ x1 * x2, data = df.tr )
The dependend variable obviously is not normally distributed, also the lognormal is not optimal so we think quasi poisson should be best.
On the training set the residuals for the quasi poisson are a lot better the the gaussian model.
But we cant seem to beat the gaussian model on a validation set.
How can this be the case? Or what are we missing?
Model summary:
GAUSSIAN
> summary(m1g)
Call:
glm(formula = y ~ x1 * x2, data = df.tr)
Deviance Residuals:
Min 1Q Median 3Q Max
-24.663 -2.849 -1.103 1.680 36.852
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.5072026 0.0841578 41.674 < 2e-16 ***
x123 -2.2156110 0.2531252 -8.753 < 2e-16 ***
x124 0.2242957 0.1762014 1.273 0.203037
x125 1.8011048 0.1166589 15.439 < 2e-16 ***
x126 -0.0409214 0.1239042 -0.330 0.741199
x127 0.8896270 0.1363625 6.524 6.86e-11 ***
x128 -0.2410489 0.1604645 -1.502 0.133048
x129 0.2261857 0.2166130 1.044 0.296397
x137 3.3886973 0.3267226 10.372 < 2e-16 ***
x139 -2.4271957 0.2900612 -8.368 < 2e-16 ***
x140 -0.6184697 0.1820797 -3.397 0.000682 ***
x143 -1.6911565 0.3578242 -4.726 2.29e-06 ***
x146 -1.8280841 0.1685329 -10.847 < 2e-16 ***
x147 0.3685382 0.1221236 3.018 0.002547 **
x148 1.1061975 0.1372421 8.060 7.66e-16 ***
x149 -0.0386354 0.1523535 -0.254 0.799812
x150 2.3230544 0.1218802 19.060 < 2e-16 ***
x151 1.3391181 0.1195007 11.206 < 2e-16 ***
x154 0.9155074 0.1156130 7.919 2.41e-15 ***
x155 -0.4611950 0.1148722 -4.015 5.95e-05 ***
x156 -0.1067856 0.1132884 -0.943 0.345887
x157 -0.8609612 0.1216078 -7.080 1.45e-12 ***
x158 -0.6485214 0.1169027 -5.548 2.90e-08 ***
x159 -0.1946808 0.1168510 -1.666 0.095703 .
x160 2.2581502 0.1283607 17.592 < 2e-16 ***
x190 -1.5436128 0.1248215 -12.367 < 2e-16 ***
x191 0.6596575 0.1149799 5.737 9.64e-09 ***
x192 0.1359673 0.1135766 1.197 0.231253
x193 -0.1363663 0.1159813 -1.176 0.239692
x194 0.5937715 0.1136418 5.225 1.74e-07 ***
x195 -0.7601841 0.1283618 -5.922 3.18e-09 ***
x196 -1.3666622 0.1251636 -10.919 < 2e-16 ***
x197 0.2268601 0.1251627 1.813 0.069907 .
x2 0.0438423 0.0005247 83.564 < 2e-16 ***
x123:x2 0.0125207 0.0021183 5.911 3.41e-09 ***
x124:x2 0.0059799 0.0014012 4.268 1.98e-05 ***
x125:x2 -0.0051185 0.0006655 -7.692 1.46e-14 ***
x126:x2 0.0020976 0.0007717 2.718 0.006568 **
x127:x2 -0.0016735 0.0009182 -1.823 0.068367 .
x128:x2 -0.0003740 0.0012000 -0.312 0.755268
x129:x2 0.0045784 0.0019535 2.344 0.019096 *
x137:x2 -0.0161315 0.0017495 -9.220 < 2e-16 ***
x139:x2 0.0325676 0.0039780 8.187 2.69e-16 ***
x140:x2 0.0007374 0.0016545 0.446 0.655808
x143:x2 -0.0071728 0.0044617 -1.608 0.107917
x146:x2 0.0115894 0.0012681 9.139 < 2e-16 ***
x147:x2 0.0049920 0.0007561 6.602 4.06e-11 ***
x148:x2 0.0007417 0.0008443 0.879 0.379668
x149:x2 0.0159149 0.0008658 18.383 < 2e-16 ***
x150:x2 -0.0011305 0.0006279 -1.800 0.071813 .
x151:x2 -0.0018370 0.0006683 -2.749 0.005980 **
x154:x2 -0.0090907 0.0007118 -12.771 < 2e-16 ***
x155:x2 -0.0025856 0.0006574 -3.933 8.38e-05 ***
x156:x2 -0.0064424 0.0006228 -10.344 < 2e-16 ***
x157:x2 0.0022582 0.0006704 3.368 0.000756 ***
x158:x2 -0.0012893 0.0006624 -1.946 0.051619 .
x159:x2 -0.0029377 0.0006449 -4.555 5.24e-06 ***
x160:x2 -0.0027016 0.0006721 -4.020 5.82e-05 ***
x190:x2 0.0070651 0.0007176 9.845 < 2e-16 ***
x191:x2 0.0039843 0.0006248 6.377 1.81e-10 ***
x192:x2 0.0007105 0.0006126 1.160 0.246164
x193:x2 0.0053882 0.0006294 8.561 < 2e-16 ***
x194:x2 0.0092873 0.0006285 14.777 < 2e-16 ***
x195:x2 0.0068949 0.0007203 9.572 < 2e-16 ***
x196:x2 0.0211705 0.0006963 30.402 < 2e-16 ***
x197:x2 0.0085271 0.0007383 11.550 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 24.77175)
Null deviance: 13114523 on 217221 degrees of freedom
Residual deviance: 5379335 on 217156 degrees of freedom
AIC: 1313736
Number of Fisher Scoring iterations: 2
QUASI POISSON
> summary(m1qp)
Call:
glm(formula = y ~ x1 * x2, family = "quasipoisson", data = df.tr)
Deviance Residuals:
Min 1Q Median 3Q Max
-7.1160 -1.1881 -0.3811 0.6225 8.5814
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.620e+00 1.021e-02 158.706 < 2e-16 ***
x123 -5.363e-01 3.787e-02 -14.160 < 2e-16 ***
x124 -2.613e-02 2.186e-02 -1.195 0.232091
x125 3.103e-01 1.335e-02 23.245 < 2e-16 ***
x126 4.703e-03 1.505e-02 0.312 0.754671
x127 9.312e-02 1.637e-02 5.689 1.28e-08 ***
x128 -9.756e-02 2.004e-02 -4.867 1.13e-06 ***
x129 -5.968e-02 2.739e-02 -2.179 0.029366 *
x137 3.954e-01 3.645e-02 10.847 < 2e-16 ***
x139 -7.190e-01 4.710e-02 -15.266 < 2e-16 ***
x140 -3.025e-01 2.572e-02 -11.759 < 2e-16 ***
x143 -7.921e-01 6.613e-02 -11.977 < 2e-16 ***
x146 -4.221e-01 2.343e-02 -18.012 < 2e-16 ***
x147 8.399e-02 1.460e-02 5.754 8.72e-09 ***
x148 1.914e-01 1.596e-02 11.995 < 2e-16 ***
x149 1.806e-01 1.723e-02 10.477 < 2e-16 ***
x150 4.901e-01 1.326e-02 36.947 < 2e-16 ***
x151 2.632e-01 1.371e-02 19.192 < 2e-16 ***
x154 7.562e-02 1.403e-02 5.388 7.13e-08 ***
x155 -5.266e-02 1.409e-02 -3.738 0.000186 ***
x156 1.221e-03 1.384e-02 0.088 0.929705
x157 -2.899e-02 1.459e-02 -1.987 0.046921 *
x158 -5.871e-02 1.426e-02 -4.118 3.82e-05 ***
x159 3.489e-02 1.400e-02 2.492 0.012706 *
x160 3.945e-01 1.430e-02 27.577 < 2e-16 ***
x190 -1.491e-01 1.547e-02 -9.635 < 2e-16 ***
x191 3.375e-01 1.287e-02 26.220 < 2e-16 ***
x192 1.980e-01 1.316e-02 15.040 < 2e-16 ***
x193 2.456e-01 1.307e-02 18.785 < 2e-16 ***
x194 3.872e-01 1.257e-02 30.813 < 2e-16 ***
x195 -1.913e-02 1.559e-02 -1.227 0.219708
x196 8.074e-02 1.446e-02 5.584 2.36e-08 ***
x197 1.808e-01 1.430e-02 12.644 < 2e-16 ***
x2 3.896e-03 4.775e-05 81.584 < 2e-16 ***
x123:x2 2.894e-03 2.228e-04 12.986 < 2e-16 ***
x124:x2 1.056e-03 1.363e-04 7.746 9.50e-15 ***
x125:x2 -1.201e-03 5.768e-05 -20.823 < 2e-16 ***
x126:x2 7.827e-05 6.976e-05 1.122 0.261896
x127:x2 -2.167e-05 8.574e-05 -0.253 0.800490
x128:x2 4.151e-04 1.111e-04 3.736 0.000187 ***
x129:x2 1.283e-03 1.974e-04 6.497 8.23e-11 ***
x137:x2 -1.570e-03 1.652e-04 -9.505 < 2e-16 ***
x139:x2 7.379e-03 4.674e-04 15.787 < 2e-16 ***
x140:x2 1.995e-03 1.870e-04 10.667 < 2e-16 ***
x143:x2 4.490e-03 7.130e-04 6.297 3.03e-10 ***
x146:x2 2.632e-03 1.355e-04 19.427 < 2e-16 ***
x147:x2 1.392e-04 6.840e-05 2.036 0.041782 *
x148:x2 -4.827e-04 7.295e-05 -6.618 3.66e-11 ***
x149:x2 -2.145e-05 7.155e-05 -0.300 0.764367
x150:x2 -1.617e-03 5.328e-05 -30.347 < 2e-16 ***
x151:x2 -8.422e-04 5.930e-05 -14.202 < 2e-16 ***
x154:x2 -5.533e-04 6.708e-05 -8.249 < 2e-16 ***
x155:x2 -2.465e-04 6.040e-05 -4.081 4.48e-05 ***
x156:x2 -6.778e-04 5.719e-05 -11.852 < 2e-16 ***
x157:x2 -1.099e-04 6.114e-05 -1.797 0.072403 .
x158:x2 -1.491e-04 6.112e-05 -2.439 0.014727 *
x159:x2 -5.313e-04 5.873e-05 -9.047 < 2e-16 ***
x160:x2 -1.210e-03 5.871e-05 -20.613 < 2e-16 ***
x190:x2 3.115e-04 6.433e-05 4.842 1.28e-06 ***
x191:x2 -1.283e-03 5.282e-05 -24.281 < 2e-16 ***
x192:x2 -1.045e-03 5.344e-05 -19.559 < 2e-16 ***
x193:x2 -1.033e-03 5.350e-05 -19.309 < 2e-16 ***
x194:x2 -1.242e-03 5.235e-05 -23.719 < 2e-16 ***
x195:x2 7.185e-05 6.448e-05 1.114 0.265129
x196:x2 2.888e-04 5.990e-05 4.821 1.43e-06 ***
x197:x2 -3.369e-04 6.226e-05 -5.411 6.29e-08 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for quasipoisson family taken to be 2.80759)
Null deviance: 1145349 on 217221 degrees of freedom
Residual deviance: 538764 on 217156 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 5