I am trying to interpret the outcome of a test for assumption of linearity. This is the dataframe:
df <- structure(list(treatment = structure(c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("CCF",
"UN"), class = "factor"), random1 = structure(c(3L, 1L, 2L, 2L,
2L, 2L, 2L, 4L, 4L, 4L, 4L, 3L, 4L, 3L, 3L, 3L, 1L, 1L, 1L, 1L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L), .Label = c("1.6",
"2", "3.2", "5", NA), class = "factor"), random2 = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L,
2L, 2L), .Label = c("1", "2", "3", "4", "5", "NA"), class = "factor"),
continuous = c(13.4834098739318, 13.5656429433723, 12.4635727711507,
18.72345150947, 18.4616104687818, 20.5685002028439, 13.8419704601596,
16.1418346212744, 17.2712407613484, 15.6206999481025, 17.3198253734436,
15.9326515550379, 13.6664227787624, 18.4006445221394, 15.9590212502841,
18.8509698995243, 20.5492911251772, 12.0971869009945, 14.2687663092537,
17.5558622926168, 12.0655307162184, 20.0060355952652, 15.9836412635937,
18.5999712367426, 14.9125382681618, 18.4091462029293, 18.766029822543,
15.8768079929326, 14.5894782578156, 11.6426318894049, 16.8206949611527,
17.0666712246649, 16.7071675430987, 16.2745705651548, 15.9203707655043
)), class = "data.frame", row.names = c(3L, 6L, 9L, 12L,
15L, 18L, 21L, 24L, 27L, 30L, 33L, 36L, 39L, 42L, 45L, 48L, 51L,
54L, 57L, 60L, 63L, 66L, 69L, 72L, 75L, 78L, 81L, 84L, 87L, 90L,
93L, 96L, 99L, 102L, 105L))
This in the model:
library(lme4)
model <- lmer((continuous) ~ treatment +(1|random1) + (1|random2), data= df, REML = TRUE)
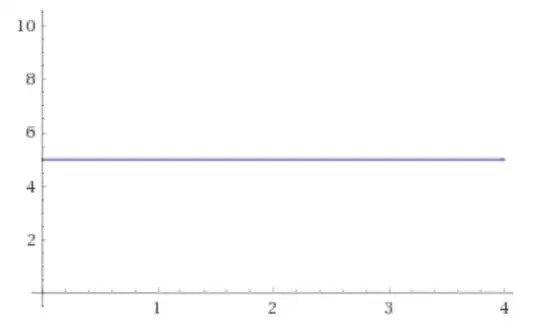
Upon checking the relationship between the independent and dependent variables to be linear with Linearity<-plot(resid(model),df$continuous) I got this result:
I was expecting my results to be either randomly scattered (linear relationship) or to show other behaviors (e.g. curvilinear relationship). How should I interpret this outcome almost completely on a straight line, and does it meet the linearity assumption?
This is what I get when I use Linearity<-plot(resid(model), fitted(model)):
This is the boxplot by treatments (colours are random1)
emmeans model results displaying estimated marginal means +- SE:
Example of random1 eeffect on CCF (different variable, see comments):