I am testing the relative odds of two groups, placebo vs active treatment, guessing that they received active treatment. These guesses were made at four time points, 4, 8, 12, and 24 weeks into a clinical trial.
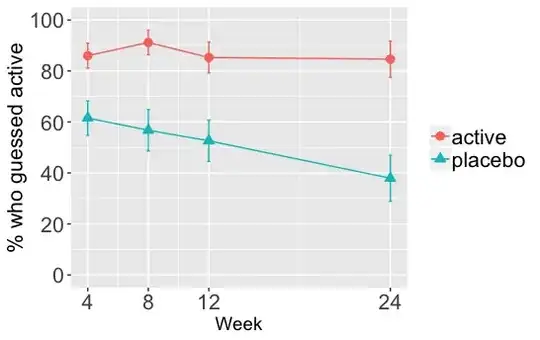
This is a graph of the percentage of people in each group who guessed they received active treatment over time
When I perform a series of logistic regressions, regressing guessed treatment on actual treatment at each of these time points in isolation, using this sort of syntax
glm(guess ~ group, data = df, family = binomial, subset = week == 4)
I get these coefficients and odds ratios
week coef or
1 4 1.345286 3.839286
2 8 2.063441 7.873016
3 12 1.652497 5.220000
4 24 2.197225 9.000000
Consistent with the graph, the odds of a person guessing that they received the active treatment are consistently higher in the active group than the placebo group (a failure of blinding basically). The odds ratio starts at around 4 on week 4 but by week 24 the odds ratio is 9.
When I perform a longitudinal logistic regression, with participant id as the random factor, and using simple coding so that the estimates for each factor, group and week (the time factor), represent the effect of that factor on log-odds averaged across all levels of the other factor (see here), like this...
# write function for creating simple coding matrix
simpMatFunct <- function (nLevels) {
k <- nLevels
mat <- matrix(rep(-1/k, k*(k-1)), ncol = k-1) + contr.treatment(k)
return(mat)
}
# run the glmer, creating simple coding matrix for each factor by using the matrix function above
glmer(guess ~ group*week + (1|id),
data = w24,
family = binomial,
contrasts = list(group = simpMatFunct(2), week = simpMatFunct(4)),
control = glmerControl(optimizer = "bobyqa"))
...these are the coefficients
Fixed Effects:
(Intercept) group2 weekFac2 weekFac3
10.2474 5.0411 2.8542 -1.8699
weekFac4 group2:weekFac2 group2:weekFac3 group2:weekFac4
0.7396 7.8657 0.8067 9.5187
The group2 coefficient, which represents the between-group difference in odds of participants guessing they recieved the active treatment, averaged across all time points, when exponentiated..
exp(5.0411)
[1] 154.64
yields an odds ratio of 150!!
Could this estimate be correct? Does the cumulative effect of consistent smaller odds ratios over time translate to a much larger odds ratio when all time points are considered together? or am I doing something wrong?