Well, we start with reading in the data in R and computing per mille (more natural with this data than %):
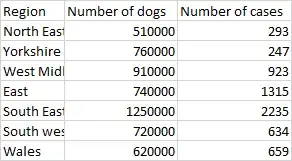
dogs
Region dogs homes
1 North E 510000 293
2 Yorkshire 760000 247
3 West Mid 910000 923
4 East 740000 1315
5 South E 1250000 2235
6 South W 720000 634
7 Wales 620000 659
pmil <- 1000*dogs$homes/dogs$dogs
paste(round(pmil, 2), "\u2030", sep="")
[1] "0.57‰" "0.32‰" "1.01‰" "1.78‰" "1.79‰" "0.88‰" "1.06‰"
So there is clearly differences. To test it we can just use the chisquare test. But then first we must get the data in the format of a contingency table (no double counting)
dogs.cont <- dogs
dogs.cont$dogs <- dogs.cont$dogs-dogs.cont$homes
chisq.test(dogs.cont[, 2:3])
Pearson's Chi-squared test
data: dogs.cont[, 2:3]
X-squared = 1364.2, df = 6, p-value < 2.2e-16
We can also compute confidence intervals (which is assuming binomial distribution) by
conf <- Hmisc::binconf(dogs$homes, dogs$dogs)
rownames(conf) <- dogs[, 1]
conf
PointEst Lower Upper
North E 0.0005745098 0.0005124005 0.0006441426
Yorkshire 0.0003250000 0.0002869231 0.0003681282
West Mid 0.0010142857 0.0009509569 0.0010818273
East 0.0017770270 0.0016836182 0.0018756086
South E 0.0017880000 0.0017154546 0.0018636076
South W 0.0008805556 0.0008146546 0.0009517824
Wales 0.0010629032 0.0009848271 0.0011471620
which we will show by plotting:
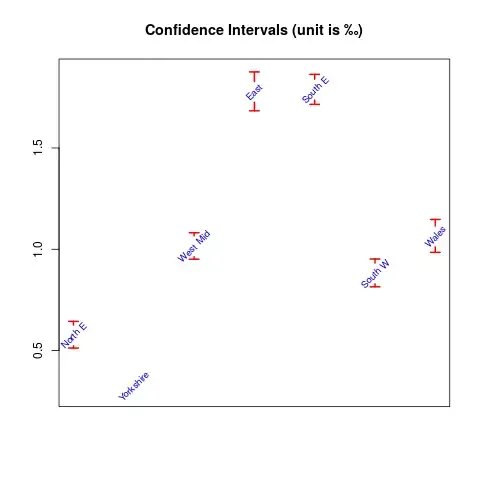
The R code used was:
conf <- 1000*conf
plotrix::plotCI(1:7, conf[, "PointEst"], li=conf[, "Lower"],
ui=conf[, "Upper"], gap=0.03, col="red", lwd=2,
main="Confidence Intervals (unit is \u2030)", xaxt="n",
xlab="", ylab="", pch=NA)
text(1:7, conf[, "PointEst"], rownames(conf), cex=0.8,
col="blue", srt=45)
The data frame can be loaded into R from:
dput(dogs)
structure(list(Region = structure(c(2L, 7L, 6L, 1L, 3L, 4L,
5L), .Label = c("East", "North E", "South E", "South W",
"Wales", "West Mid", "Yorkshire"), class = "factor"),
dogs = c(510000, 760000, 910000, 740000, 1250000, 720000,
620000), homes = c(293, 247, 923, 1315, 2235, 634, 659)),
class = "data.frame", row.names = c(NA, -7L))