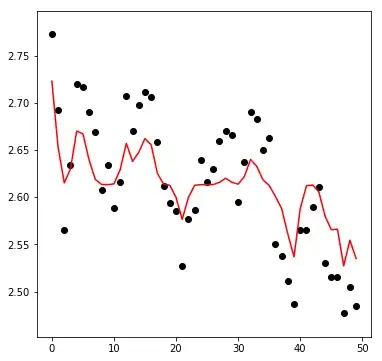
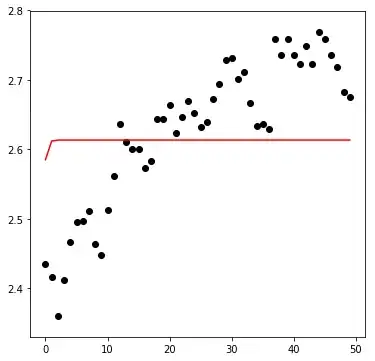
What is observed is a direct consequence of using a potentially improper parameter set for svr. It appears that especially the epsilon used, i.e. the minimum distance between actual and predicted values that needs to be observed so a penalty is induced during fitting, is somewhat large; therefore errors are not adequately penalised and this leads to potential under-fitting. In addition as there is an obvious sequential nature to our data, the influence of the parameter gamma, i.e. how far the influence of a single training example reaches, is going to be important too.
I quickly eye-balled the data from the graph shown and made some quick predictions to show how strong the effect of epsilon can be in this case.
Some final application-specific comments:
- The flat estimate is not necessarily bad! Off the bat I would say it is actually "correct"; following Makrydakis et al. (2018) Statistical and Machine Learning forecasting methods: Concerns and ways forward setting
epsilon "equal to the noise level of the training sample" is a perfectly reasonable thing to start with. "Beating the historical mean" as a forecast (as the current flat-line suggests), is not as trivial as it sounds (see the excellent CV thread on : Is it unusual for the MEAN to outperform ARIMA? for more details).
- Time-series forecasting tasks tend to be dominated by exponential smoothing and/or ARIMA models; SVMs, while promising, never really established themselves as a strong alternative. I would suggest you consider such approaches if you have not done already.
- Consider using rolling time-window approach when cross-validating the fitting procedure for a time-series model. Rob Hyndman has written extensively on time-series cross-validation, his book with Athanasopoulos has a very nice section about this available here. In Python check
TimeSeriesSplit from sklearn.model_selection.
Code:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.svm import SVR
dtrain=np.random.uniform(0,0.00100,[100,2])
dtrain[:50,1] = np.array([2.770, 2.690, 2.560, 2.630, 2.730, 2.720, 2.690, 2.680, 2.600, 2.630,
2.580, 2.615, 2.72, 2.670, 2.690, 2.710, 2.705, 2.660, 2.610, 2.580,
2.650, 2.600, 2.550, 2.600, 2.630, 2.61, 2.620, 2.660, 2.670, 2.665,
2.595, 2.625, 2.685, 2.680, 2.650, 2.660, 2.550, 2.540, 2.520, 2.486,
2.551, 2.551, 2.580, 2.605, 2.525, 2.510, 2.510, 2.480, 2.515, 2.485])
dtrain[:,0] = np.arange(1,101)
X = dtrain[:50,0]
y = dtrain[:50,1]
X_test = dtrain[50:99,0]
svr_rbf_original = SVR(kernel='rbf', epsilon=0.05)
svr_rbf_original.fit(X.reshape(-1,1),y)
svr_rbf_twicked = SVR(kernel='rbf', epsilon=0.003, gamma=0.001)
svr_rbf_twicked.fit(X.reshape(-1,1),y)
plt.plot(X, y,linestyle="",marker="o", label='Raw')
plt.plot(X, svr_rbf_original.predict(X.reshape(-1,1)), label='Within sample fit')
plt.plot(X_test, svr_rbf_original.predict(X_test.reshape(-1,1)), label='Fit eps:0.05')
plt.plot(X_test, svr_rbf_twicked.predict(X_test.reshape(-1,1)), label='Fit eps:0.003 / gamma:0.001')
plt.legend()
plt.show()