I have a dataset associated with labels. According to https://arxiv.org/pdf/1802.03426.pdf --> UMAP (Uniform Manifold Approximation and Projection) which is a novel manifold learning technique for dimension reduction and the data, I succeeded to create the green and red clouds bellow. The problem I have is they are stick together. For machine learning purposes, it is kinda hard to learn something when the clouds are placed that way.
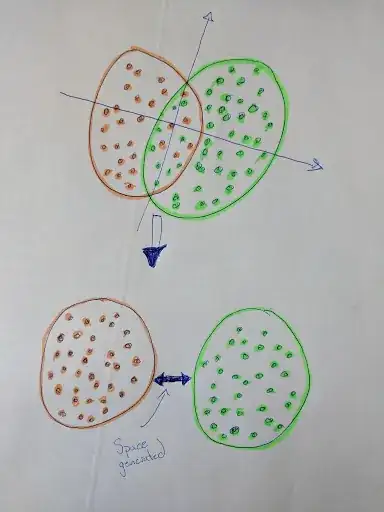
Is there a topological approach that might be used to create a significant space between clouds?
UPDATE
I would be interested by an analytic approach to separated the two clouds. Each cloud can be seen as a compact space.
Here is an example in 2-D. I would like a way to generalize that concept in z-D, where z would be a finite positive integer.
UPDATE
I have already used several approaches to tackle my problem, i.e. PCA, t-SNE, SVM, using a neural network to classify the points. The problem is I got always two clouds but stick together. UMAP gave me the best results. Now, once UMAP has been applied, I want a way to force the clouds to be separated in knowing the clouds are already well distinct.
UPDATE 2
I am trying an approach, but I am far from certain that it's the best solution.
- Cover each cloud by the smallest possible sphere.
- Extend the intersection of the spheres by an hyperplane.
- Taking away the clouds according to the orthogonal vector to the hyperplane by a distance alpha.
UPDATE 3
def umap_plot(trans, label, transform=False):
fig = plt.figure(figsize = (8,8))
ax = fig.add_subplot(1, 1, 1, projection='3d')
ax.set_title('UMAP - 3D representation', fontsize = 20)
targets = [1, -1]
colors = ['g', 'r']
for target, color in zip(targets, colors):
indicesToKeep = np.array([i==target for i in label])
if target == 1 and transform == True:
trans.embedding_[indicesToKeep] += 20
ax.scatter(trans.embedding_[indicesToKeep][:, 0],
trans.embedding_[indicesToKeep][:, 1],
trans.embedding_[indicesToKeep][:, 2],
c = color,
s = 20)
ax.legend(targets)
ax.grid()
plt.show()
from random import shuffle
listing = list(zip(trans.embedding_.tolist(), y_train))
shuffle(listing)
trans.embedding_, y_train = list(zip(*listing))
trans.embedding_, y_train = np.array(trans.embedding_), np.array(y_train)
umap_plot(trans, label)
Even shuffling the data gave me the same plot result