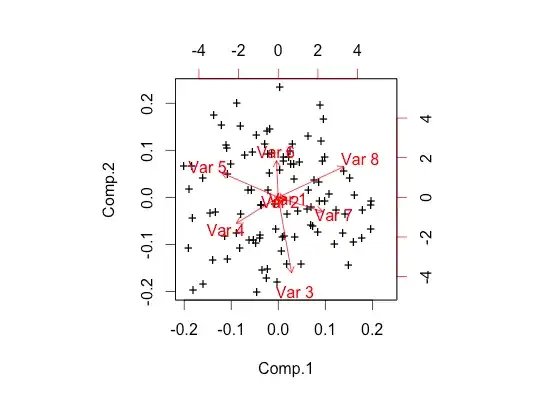
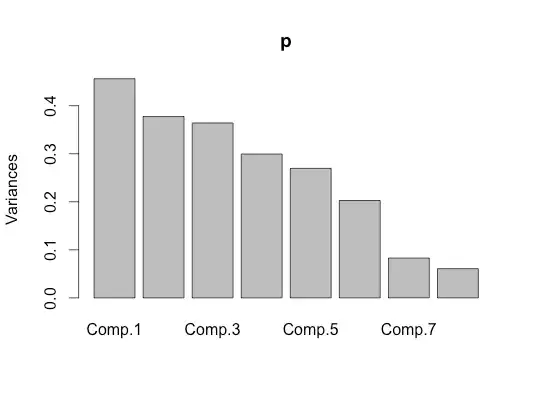
I just wanted to do this small experiment to make sure I understand PCA correctly. My dataset contains 8 columns. The first two columns are randomly generated in excel => randbetween(4, 5) and the other 6 columns are also generated in the same way but the formula used is => randbetween(1,3)
When I do PCA on this I am not getting good results. I expect that the result should indicate high eigenvalues for a factor that is a combination of first two columns and low on other colums. This is my code in R :
sensex.dat = read.csv('C:/Study/_SEM4/brand man/emperical/dice.csv', header = T)
attach(sensex.dat)
sensex.cov = cov(sensex.dat)
sensex.eigen = eigen(sensex.cov, symmetric = T)
sensex.eigen$values
sensex.eigen$vectors