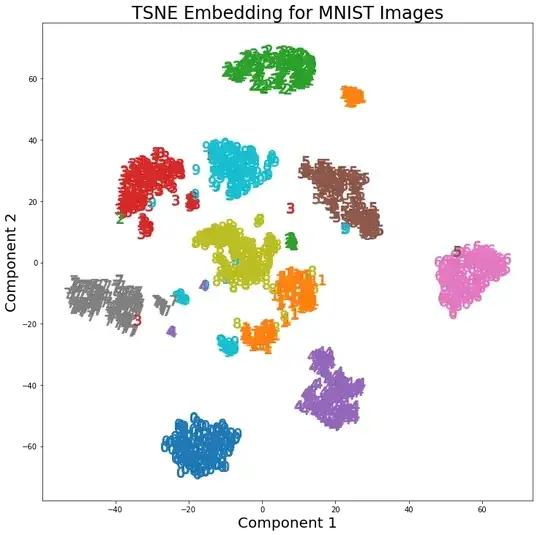
In a recent assignment, we were told to use PCA on the MNIST digits to reduce the dimensions from 64 (8 x 8 images) to 2. We then had to cluster the digits using a Gaussian Mixture Model. PCA using only 2 principal components does not yield distinct clusters and as a result the model is not able to produce useful groupings.
However, using t-SNE with 2 components, the clusters are much better separated. The Gaussian Mixture Model produces more distinct clusters when applied to the t-SNE components.
The difference in PCA with 2 components and t-SNE with 2 components can be seen in the following pair of images where the transformations have been applied to the MNIST dataset.
I have read that t-SNE is only used for visualization of high dimensional data, such as in this answer, yet given the distinct clusters it produces, why is it not used as a dimensionality reduction technique that is then used for classification models or as a standalone clustering method?